Abstract
Background
There has been uncertainty about the safety or benefit of angiotensin-converting enzyme (ACE) inhibitors during the COVID-19 pandemic. We used Mendelian randomization using genetic determinants of serum-ACE levels to test whether decreased ACE levels increase susceptibility to SARS-CoV-2 infection or COVID-19 severity, while reducing potential bias from confounding and reverse causation in observational studies.
Methods
Genetic variants strongly associated with ACE levels, which were nearby the ACE gene, were identified from the ORIGIN trial and a separate genome-wide association study (GWAS) of ACE levels from the AGES cohort. The ORIGIN trial included 4147 individuals of European and Latino ancestries. Sensitivity analyses were performed using a study of 3200 Icelanders. Cohorts from the COVID-19 Host Genetics Initiative GWAS of up to 960 186 individuals of European ancestry were used for COVID-19 susceptibility, hospitalization and severe-disease outcome.
Results
Genetic variants were identified that explain between 18% and 37% of variance in ACE levels. Using genetic variants from the ORIGIN trial, a standard-deviation decrease in ACE levels was not associated with an increase in COVID-19 susceptibility [odds ratio (OR): 1.02, 95% confidence interval (CI): 0.90, 1.15], hospitalization (OR: 0.86, 95% CI: 0.68, 1.08) or severe disease (OR: 0.74, 95% CI: 0.51, 1.06). Using genetic variants from the AGES cohort, the result was similar for susceptibility (OR: 0.98, 95% CI: 0.89, 1.09), hospitalization (OR: 0.86, 95% CI: 0.66, 1.11) and severity (OR: 0.75, 95% CI: 0.50, 1.14). Multiple-sensitivity analyses led to similar results.
Conclusion
Genetically decreased serum ACE levels were not associated with susceptibility to, or severity of, COVID-19 disease. These data suggest that individuals taking ACE inhibitors should not discontinue therapy during the COVID-19 pandemic.
Keywords: COVID-19, angiotensin-converting enzyme, ACE inhibitors, Mendelian randomization
Key Messages
The SARS-CoV-2 virus uses the angiotensin-converting enzyme 2 (ACE2) to invade host cells.
ACE2 is a close analogue to the angiotensin-converting enzyme (ACE) and they are both involved in the renin–angiotensin–aldosterone system. It is unclear how ACE inhibitors may affect ACE2 regulation, but these medications have been speculated to lead to a compensatory increase in ACE2, thus potentially increasing susceptibility to, and severity of, COVID-19.
There is clinical equipoise as to the harm or benefit of ACE inhibitors during the COVID-19 pandemic and available evidence is retrospective, observational and at high risk of confounding and reverse-causation bias.
The use of genetic variants associated with lowered ACE levels through Mendelian randomization can provide insight into the effect of ACE inhibition on COVID-19 outcomes, while avoiding bias due to confounding and reverse causation.
Lowered ACE levels were not associated with increased susceptibility to, or severity of, COVID-19.
Introduction
As the cause of the ongoing COVID-19 pandemic, SARS-CoV-2 invades host cells by attaching to the membrane-bound angiotensin-converting enzyme 2 (ACE2).1 ACE2 shares similarities with its protein homolog angiotensin-converting enzyme (ACE) and both play a role in the renin–angiotensin–aldosterone system. However, ACE2 differs in substrate and tissue expression, and, importantly, ACE inhibitors do not inhibit ACE2.2 ACE inhibitors are a class of antihypertensive agents with benefits in many common cardiovascular diseases3 and are prescribed to >21% of adults aged 60–79 in the USA and Canada.4 Despite conflicting supporting in-vivo evidence, their shared metabolic pathway has led to concerns over the use of ACE inhibitors during the COVID-19 pandemic.5 Specifically, if ACE inhibitors lead to decreased ACE levels, causing a compensatory ACE2 upregulation, then a large proportion of the population could be at an increased risk of COVID-19 due to the use of ACE inhibitors. Whereas some retrospective studies did not show evidence of harm from ACE inhibition,6–8 they may have been underpowered. For example, using a cohort from Denmark, Fosbøl et al.6 found a 30-day-mortality hazards ratio (HR) of 0.83 [95% confidence interval (CI): 0.67–1.03] in those not on ACE inhibitors and no effect on COVID-19 susceptibility (HR: 1.05, 95% CI: 0.80, 1.36). Moreover, other observational studies have shown benefits in ACE inhibitors, or even suggested a biphasic effect of ACE inhibitors,9 depending on the stage of COVID-19.
Given the clinical equipoise and a scarcity of data, most medical societies have opted for a ‘first do no harm’ approach, recommending not to modify ACE-inhibitor therapy to prevent COVID-19 complications until better data are available.10,11 Unfortunately, given the clear benefits of ACE inhibitors in many diseases, randomized prospective trials are likely to suffer from indication bias, in which patients with the greatest risk for severe COVID-19 are also those with the greatest need for an ACE inhibitor, and therefore are unlikely to be enrolled in an ACE-inhibitor trial. Moreover, current observational epidemiological studies that estimated the effect of ACE inhibitors on COVID-19 were likely subject to confounding and reverse causation.12,13 Confounding happens when ACE-inhibitor prescription and COVID-19 are influenced by a third variable (such as cardiovascular diseases), which is not in the causal pathway between them. Reverse causation may also bias such studies. This bias occurs when the outcome influences the exposure. Even with sophisticated statistical adjustments, traditional epidemiological studies are therefore at risk of providing biased estimates of the causal effect of ACE inhibitors on COVID-19.
One way to reduce risk of both biases is Mendelian randomization (MR)—a genetic epidemiology method that uses genetic determinants of the exposure (ACE level) to understand the effect of the exposure on the outcome (COVID-19 susceptibility and severity). Since genetic variants are randomly assigned at conception, this breaks the association with nearly all confounding factors. Also, genetic variants are always assigned prior to disease onset, thereby precluding reverse causation.14 MR has three main assumptions.15 First, the genetic variants must be associated with the exposure (here, serum ACE levels). Second, the genetic variants must not be associated with confounders of the relationship between the exposure and the outcome (here, COVID-19 susceptibility and severity), e.g. through population stratification. Lastly, the variants must only be associated with the outcome of interest through their effect on the exposure (also known as an absence of horizontal pleiotropy).16
ACE inhibitors act by decreasing ACE activity. Given that ACE activity is mediated in part by circulating ACE levels,17,18 by selecting genetic variants associated with lower serum ACE levels, we can provide insights into the effect of ACE inhibitors on susceptibility to, and severity of, COVID-19. In this study, we use such variants as genetic instruments for the effect of decreased ACE levels on COVID-19 susceptibility and severity as part of an MR study. This approach thereby can provide estimates of the effect of ACE inhibitors while reducing bias due to confounding and reverse causation.
Methods
Study design
We performed a two-sample MR analysis to study the effect of ACE-serum levels on COVID-19 susceptibility and severity. This method measures the effect of genetic variants on ACE levels and COVID-19 using separate data sets for the exposure and outcome, allowing increased sample size and statistical power, while lowering bias from weak genetic instruments.19 These data sources are summarized in Table 1 and Supplementary Table 1, available as Supplementary data at IJE online.
Table 1.
Sources of data for the analysis
| Phenotype | Source of genetic variants |
|
|---|---|---|
| Consortium | Participants | |
| Serum-ACE levels | ORIGIN trial genetic and biomarker substudies20,21 |
A sub-study of patients originally enrolled to the ORIGIN trial who had both whole-genome genotyping and serum-ACE levels measured Basic demographics for the genetic sub-study20:
|
| AGES Reykjavik study22 | Genome-wide association study of ACE circulating level in 3200 Icelanders over the age of 65. Basic demographics:
|
|
| COVID-19 susceptibility | Susceptibility |
Cases: 3382 individuals with COVID-19 by laboratory confirmation, chart review or self-report Controls: 37 851 individuals without COVID-19 by laboratory confirmation or self-report |
| Extended susceptibility |
Cases: 6182 individuals with COVID-19 by laboratory confirmation, chart review or self-report Controls: 960 186 individuals enrolled in the cohorts and not included as cases |
|
| COVID-19 severity | Hospitalized |
Cases: 677 hospitalized individuals with COVID-19 Controls: 2372 non-hospitalized individuals with COVID-19 |
| Extended hospitalized |
Cases: 2710 hospitalized individuals with COVID-19 Controls: 813 234 individuals enrolled in the cohorts and not included as cases |
|
| Severe disease |
Cases: 213 COVID-19-infected hospitalized individuals who died or required respiratory support (intubation, CPAP, BiPAP, continuous external negative pressure, high-flow nasal cannula) Controls: 750 non-hospitalized individuals with COVID-19 |
|
| Extended severe disease |
Cases: 540 COVID-19-infected hospitalized individuals who died or required respiratory support (intubation, CPAP, BiPAP, continuous external negative pressure, high-flow nasal cannula) Controls: 366 840 individuals enrolled in the cohorts and not included as cases |
|
See Supplementary Table 1, available as Supplementary data at IJE online, for details on cohorts of COVID-19 susceptibility and severity phenotypes. ACE, angiotensin-converting enzyme.
ACE genetic variants data source
Our choice of the genetic variants associated with ACE-serum levels is based on Pigeyre et al.’s20 MR study, in which these genetic variants were used to show that a lowered ACE-serum level decreases the risk of diabetes mellitus. Briefly, the ORIGIN cohort genetic and biomarker sub-studies were used to obtain genome-wide genotyping and serum-ACE-levels measurements.20,21 Genotyping was performed on 4147 participants (including 63.25% who reported using ACE inhibitors20) using the HumanCore Exome chip (Illumina, San Diego, USA) and the 1000 Genomes Project reference panel23 was used for genotype imputation. Serum-ACE levels were quantified in 8401 ORIGIN trial participants using the Luminex 100/200 System (Luminex, Austin, USA). Details on genotype-quality control and ACE measurement can be found elsewhere.20 Linear additive genetic association was performed separately on individuals of European and Latin American ancestry using age, sex and the first five genetic principal components as covariates. Results were then meta-analysed results across ancestries to obtain genetic variants for the MR.
In Pigeyre et al.,20 genetic variants associated with serum-ACE levels were selected to be cis-acting protein quantitative trait locus (cis-pQTL) single nucleotide polymorphisms (SNPs), which were defined as being within 300 kilo-bases of the ACE gene locus (17q23.3) and pruned for linkage disequilibrium at an r2 coefficient of correlation of <10%. The use of cis-acting pQTL SNPs reduces the risk of horizontal pleiotropy, since (compared with trans variants) cis genetic variants that strongly associate with serum-ACE levels are likely to directly influence the gene’s transcription. Since this would be a direct effect, not mediated by other proteins, it reduces the probability that the selected genetic variants influence COVID-19 susceptibility and severity independently of serum-ACE levels. However, this is not always easy to demonstrate and the risk of horizontal pleiotropy should still be assessed even when using such instruments. From those, we then selected the SNPs with p < 5 × 10–8 and with minor allele frequency of >0.5% for our MR. To ensure that our results were not affected by any remaining linkage disequilibrium between genetic instruments, we also pruned SNPs to an r2 of 1%. To do so, we first selected the rs4343 SNP, given that it explained >23% of the ACE-level variance in the ORIGIN trial. Using LD Link24 and the 1000 Genome European ancestry populations, we built a linkage-disequilibrium matrix to select other SNPs associated with serum-ACE levels using an r2 of 1%.
In a sensitivity analysis (see below), we also used a previously reported cis-pQTL SNP (rs4344), previously reported in Emilsson et al.22 and identified in 3200 Icelandic individuals from the AGES Reykjavik study, since this would decrease the population bias from population stratification.
Positive control
To verify that our genetic instruments and analysis were statistically powered to detect a clinical effect of ACE levels, we used a positive control outcome anticipated to be associated with ACE levels. For this, we used a GWAS on self-reported hypertension (as a binary trait) in the UKB from the OpenGWAS project25 and accessed it through the TwoSampleMR R package (ID: UKB-b:14057). It contained 119 731 cases and 343 402 controls. However, because ACE inhibitors are used in a wide variety of highly polygenic diseases,26–28 the effect of our genetic instruments is unlikely to be large. Further, for some traits such as hypertension, using ACE inhibitors may prevent adequate trait measurement. Therefore, we used ‘being prescribed an ACE inhibitor or angiotensin-converting agent’ as an outcome for a second positive control. For this, summary statistics were obtained from a previously published GWAS documenting medication use in individuals from the UKB.29 This GWAS contained 62 752 cases and 174 778 controls.
Horizontal pleiotropy assessment
As described above, to reduce the risk of horizontal pleiotropy, all the genetic instruments from the ORIGIN trial and AGES Reykjavik study were cis-pQTL SNP. Further, we used the PhenoScanner tool30,31 to check whether any of the selected SNPs were associated with other phenotypes at risk of affecting COVID-19 susceptibility or severity independently of serum-ACE levels. To do so, we assessed SNPs at a threshold of P < 5 × 10–8 for their association with any other phenotypes. We then performed a sensitivity analysis without SNPs at high risk of horizontal pleiotropy.
COVID-19 susceptibility and severity data source
We obtained effect estimates of ACE levels on COVID-19 by obtaining effect coefficients from the above SNPs in GWAS meta-analyses from the COVID-19 Host Genetics Initiative (COVID-19 HGI).32 The COVID-19 HGI used six different case/control definitions to identify genetic variants associated with COVID-19 susceptibility and disease severity. For our study, we used a susceptibility phenotype that compared confirmed COVID-19 cases, defined as individuals with laboratory confirmation of SARS-CoV-2 infection based on nucleic acid amplification or serology based tests or by electrical health records (using International Classification of Diseases or physician notes), with controls defined as laboratory-tested negative for SARS-CoV-2 infection (for all tests if multiple were performed) or self-reported test-negative individuals (this case/control definition is labelled as C1V2 in COVID-19 HGI). Details of the UK Biobank analysis are found in Supplementary Table 4, available as Supplementary data at IJE online.
To assess COVID-19 severity, we used two approaches. First, we used a hospitalized phenotype in which cases were defined as hospitalized patients with COVID-19 and controls were COVID-19-positive non-hospitalized individuals (this case/control definition is labelled as B1V2 in COVID-19 HGI). Second, we used a severe-disease phenotype in which cases were defined as hospitalized individuals with COVID-19 and requiring respiratory support (this case/control definition is labelled as A1V2 in COVID-19 HGI). Respiratory support was defined as intubation, CPAP, BiPAP, continuous external negative pressure or high-flow nasal cannula. Controls were also non-hospitalized COVID-19-infected individuals.
Finally, we also used an extended susceptibility, an extended hospitalized and an extended severe-disease phenotype whereas controls were defined as all non-cases in the included cohorts. These case/control definitions are labelled C2V2, B2V2 and A2V2 in COVID-19 HGI, respectively. Details of the six phenotypes are found in Table 1 and Supplementary Table 1, available as Supplementary data at IJE online.
Cohorts enrolled patients and performed GWAS locally based on a standardized analysis plan and phenotype definitions. For this study, we used the individual GWASs restricted to individuals of European ancestry to reduce the risk of bias from population stratification, which we then meta-analysed using fixed-effects models with the METAL package.33
Lastly, given that the Host(a)ge cohort34 was the largest case contributor to the COVID-19 hospitalized phenotype and has already been published elsewhere, we also used this cohort’s GWAS as the outcome for a separate MR sensitivity analysis (Supplementary Table 2, available as Supplementary data at IJE online).
Positive control
To verify that our genetic instruments and analysis were statistically powered to detect a clinical effect of ACE levels, we used a positive control outcome anticipated to be associated with ACE levels. For this, we used a GWAS on self-reported hypertension (as a binary trait) in the UKB from the OpenGWAS project25 and accessed it through the TwoSampleMR R package (ID: UKB-b:14057). It contained 119 731 cases and 343 402 controls. However, because ACE inhibitors are used in a wide variety of highly polygenic diseases,26–28 the effect of our genetic instruments is unlikely to be large. Further, for some traits such as hypertension, using ACE inhibitors may prevent adequate trait measurement. Therefore, we used ‘being prescribed an ACE inhibitor or angiotensin-converting agent’ as an outcome for a second positive control. For this, summary statistics were obtained from a previously published GWAS documenting medication use in individuals from the UKB.29 This GWAS contained 62 752 cases and 174 778 controls.
Primary MR analysis
For each SNP, the SNP’s effect coefficients on serum-ACE levels and on COVID-19 susceptibility and severity were combined using the Wald ratio method to estimate the effects of ACE levels on COVID-19. Each ratio was meta-analysed using inverse-variance weighting to obtain the final effect estimate. All analyses were performed using the TwoSampleMR package35 (v0.4.25) on R (v3.5.0). Since all SNPs’ effects were estimated in all cohorts, no proxy SNPs were required for this MR analysis.
Sensitivity analysis
First, to assess whether the selected SNPs were associated with COVID-19 susceptibility and severity through a mechanism independently of serum-ACE levels (which would violate the third MR assumption), we performed MR-Egger analysis. MR-Egger performs a meta-analysis of the individual Wald ratios while allowing an additional y-intercept variable alpha. An intercept (alpha) differing from zero indicates directional horizontal pleiotropy, suggestive of a violation of the third MR assumption.
Second, to see whether our choice of r2 of <1% decreased our statistical power, we performed an analysis using the original 10% threshold from Pigeyre et al.20 We also used the PhenoScanner tool to check for SNPs at risk of pleiotropy and removed them from our MR in an additional analysis.
Finally, given that our SNPs were obtained from the ORIGIN trial, which had mixed European and Latino American ancestries (which could lead to bias from population stratification), we used a separate cis-pQTL SNP (rs4344) reported by Emilsson et al.22 as an instrument to repeat our MR analyses. This cis-pQTL SNP was obtained from the AGES Reykjavik study and we estimated that rs4344 explained 18% of the variance of ACE levels. Note that, for this sensitivity analysis, the effect estimate can only be used to infer the direction of effect, since a non-linear Yeo-Johnson transform was used on measured ACE levels.
Results
Genetic instruments
From the original 17 SNPs from Pigeyre et al.’s study, we removed 5 SNPs that did not reach a p-value threshold of p < 5 × 10–8 for their association with serum-ACE levels. All SNPs were available in all outcome-phenotype GWASs and thus no proxies were used. The final 12 genetic instruments and their summary statistics are shown in Table 2. Reasons for excluding SNPs from each analysis are given in Supplementary Table 3, available as Supplementary data at IJE online.
Table 2.
Genetic instrument summary statistics showing their effect on angiotensin-converting enzyme (ACE) levels, adapted from Pigeyre et al.20
| SNP | EA/OA | EAF—European ancestry | EAF—Latin American ancestry | Beta (s.e.) | p-value |
|---|---|---|---|---|---|
| rs4343a | A/G | 0.45 | 0.46 | −0.63 (0.02) | 1.5 × 10–213 |
| rs1074637a | T/C | 0.90 | 0.91 | −0.24 (0.04) | 4.4 × 10–09 |
| rs11650201 | G/T | 0.16 | 0.18 | −0.28 (0.03) | 2.7 × 10–18 |
| rs12452187 | A/G | 0.60 | 0.61 | −0.23 (0.02) | 2.5 × 10–27 |
| rs12602457 | G/T | 0.85 | 0.89 | −0.23 (0.03) | 2.6 × 10–14 |
| rs13342595 | C/T | 0.23 | 0.24 | −0.14 (0.02) | 2.5 × 10–09 |
| rs2137143b | T/G | 0.96 | 0.98 | −0.35 (0.06) | 7.5 × 10–09 |
| rs4968780 | C/A | 0.05 | 0.06 | −0.28 (0.05) | 1.9 × 10–08 |
| rs72847305b | A/G | 0.10 | 0.09 | −0.33 (0.04) | 4.5 × 10–17 |
| rs74251225 | G/A | 0.04 | 0.13 | −0.26 (0.04) | 1.6 × 10–10 |
| rs75457471a | A/G | 0.38 | 0.40 | −0.19 (0.02) | 8.1 × 10–15 |
| rs79480822 | C/T | 0.93 | 0.97 | −0.55 (0.05) | 6.4 × 10–24 |
SNP, single nucleotide polymorphism; EA/OA, effect allele/other allele; Beta, logistic regression coefficient of additive effect of the effect allele on ACE levels; EAF, effect allele frequency; s.e., standard error.
These SNPs were also used for the sensitivity analysis restricting to SNPs with an r2 of <1%.
SNP was deemed to be at risk of horizontal pleiotropy.
The rs4343 SNP was the variant most strongly associated with serum-ACE levels in the ORIGIN trial population and explained 21% of variance in ACE levels.20 Overall, the combination of the selected 12 SNPs explained 37% of the ACE variants, whereas the 3 SNPs used for the primary analysis retaining only SNPs with r2 of <1% (including rs4343) still explained 23% of the variance. Of note, the rs4344 SNP from the AGES Reykjavik study used for our last sensitivity analysis was in high linkage disequilibrium with rs4343 in European 1000 Genome populations (r2 = 96%).
Positive control
Using the genetic instruments from our primary analysis (r2 < 1%), a standard-deviation increase in ACE levels was associated with an increased odds of hypertension of 1.007 (95% CI: 1.004, 1.01, P = 9.6 × 10–8). Using SNPs with an r2 of <10%, we obtained an odds ratio (OR) of 1.008 (95% CI: 1.006, 1.01, P = 5.3 × 10–13). Similarly, using the primary-analysis instruments, a standard-deviation increase in ACE levels was associated with an increase in the OR of ACE-inhibitor or angiotensin-receptor-blocker use of 1.05 (95% CI: 1.02, 1.08, P = 2.4 × 10–4). Using SNPs with an r2 of <10%, we obtained an OR of 1.05 (95% CI: 1.04, 1.07, P = 3.3 × 10–10).
Cohorts used for the outcome-phenotype GWAS
The cohorts used for all the outcome phenotypes were of European ancestry. The sample size varied markedly among phenotypes (Table 1). The extended susceptibility was the largest, with 6182 cases and 960 186 controls, and the severe-disease phenotype was the smallest, with 213 cases and 750 controls. Note that the severe-disease-phenotype cases and controls were all from the UK Biobank. The cohort contributing the largest number of cases was Host(a)ge (1610 cases), but the UK Biobank contributed the largest number of individuals overall (up to 1283 cases and 364 379 controls).
Primary analysis: effect of ACE levels on COVID-19 susceptibility and severity
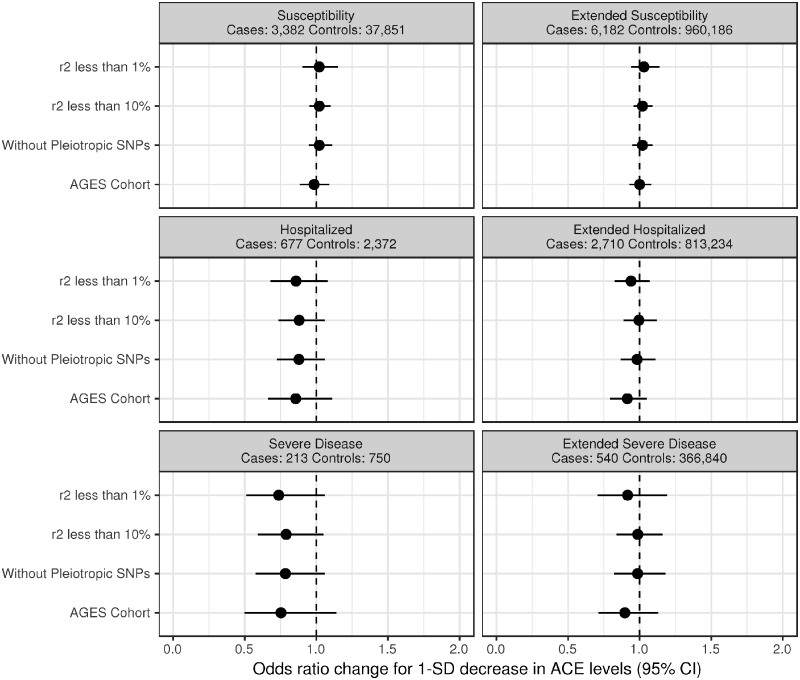
For the primary analysis (limited to SNPs having an r2 of <1%), we used the following three SNPs: rs4343, rs1074637 and rs75457471. These SNPs were not found to be at risk of pleiotropy using the PhenoScanner tool. Our MR analysis showed that a 1-standard-deviation decrease in serum-ACE levels was not associated with susceptibility (OR: 1.02, 95% CI: 0.90, 1.15, P = 0.76), extended susceptibility (OR: 1.03, 95% CI: 0.94, 1.14, P = 0.48), hospitalization (OR: 0.86, 95% CI: 0.68, 1.08, P = 0.20), extended hospitalization (OR: 0.94, 95% CI: 0.83, 1.07, P = 0.35), severe disease (OR: 0.74, 95% CI: 0.51, 1.06, P = 0.10) or extended severity (OR: 0.92, 95% CI: 0.71, 1.19, P = 0.51) (see Figure 1 and Table 3). The MR-Egger intercept term (alpha) and its 95% CIs were close to the null in all analyses, suggesting no detected evidence of directional pleiotropy.
Figure 1.
Point estimate and 95% confidence interval of a 1-standard-deviation decrease in the angiotensin-converting enzyme (ACE) level on Covid-19 susceptibility and severity. For the analysis using the AGES cohort, the estimate can only be used to infer the direction of effect. From top to bottom: primary analysis with linkage-disequilibrium coefficient (r2) <1%, sensitivity analysis with r2 <10%, sensitivity analysis with r2 <10% without single nucleotide polymorphisms (SNPs) at risk of pleiotropy and sensitivity analysis using the AGES Reykjavik cohort.
Table 3.
MR results from sensitivity analysis with linkage-disequilibrium coefficient (r2) <1%
| Inverse-variance weighted MR meta-analysis |
MR-Egger meta-analysis |
||||
|---|---|---|---|---|---|
| Odds ratio (95% CI) | p-value | Intercept | Alpha p-value | Odds ratio (95% CI) | Odds ratio p-value |
| Susceptibility | |||||
| 1.02 (0.90, 1.15) | 0.76 | –0.11 (–0.22, 0.01) | 0.33 | 0.84 (0.67, 1.07) | 0.39 |
| Extended susceptibility | |||||
| 1.03 (0.94, 1.14) | 0.48 | –0.07 (–0.18, 0.03) | 0.40 | 0.91 (0.74, 1.11) | 0.52 |
| Hospitalized | |||||
| 0.86 (0.68, 1.08) | 0.20 | 0.03 (–0.23, 0.28) | 0.86 | 0.91 (0.53, 1.54) | 0.78 |
| Extended hospitalized | |||||
| 0.94 (0.83, 1.07) | 0.35 | –0.03 (–0.28, 0.21) | 0.84 | 0.89 (0.56, 1.42) | 0.71 |
| Severe disease | |||||
| 0.74 (0.51, 1.06) | 0.10 | 0.11 (–0.27, 0.48) | 0.68 | 0.90 (0.40, 2.01) | 0.84 |
| Extended severe disease | |||||
| 0.92 (0.71, 1.19) | 0.51 | –0.008 (–0.47, 0.45) | 0.98 | 0.90 (0.36, 2.24) | 0.86 |
Odds ratios are presented for a decrease in 1 standard deviation in angiotensin-converting enzyme level. CI, confidence interval.
Sensitivity analyses
For the analysis with r2 < 10% (Table 4), a standard-deviation decrease in serum-ACE levels was also not associated with a clinically relevant change in COVID-19 susceptibility (OR: 1.02, 95% CI: 0.95, 1.10, P = 0.52), extended susceptibility (OR: 1.02, 95% CI: 0.96, 1.09, P = 0.53), hospitalization (OR: 0.88, 95% CI: 0.74, 1.06, P = 0.17), extended hospitalization (OR: 1.0, 95% CI: 0.89, 1.12, P = 0.93), severe disease (OR: 0.79, 95% CI: 0.59, 1.05, P = 0.11) or extended severe disease (OR: 0.99, 95% CI: 0.84, 1.16, P = 0.87). The MR-Egger analyses also did not suggest evidence of directional pleiotropy.
Table 4.
MR results from sensitivity analysis with linkage-disequilibrium coefficient (r2) <10%
| Inverse-variance weighted MR meta-analysis |
MR-Egger meta-analysis |
||||
|---|---|---|---|---|---|
| Odds ratio (95% CI) | p-value | Intercept | Alpha p-value | Odds ratio (95% CI) | Odds ratio p-value |
| Susceptibility | |||||
| 1.02 (0.95, 1.10) | 0.52 | –0.02 (–0.07, 0.04) | 0.60 | 0.99 (0.84, 1.15) | 0.85 |
| Extended susceptibility | |||||
| 1.02 (0.96, 1.09) | 0.53 | 0.01 (–0.04, 0.06) | 0.77 | 1.04 (0.90, 1.20) | 0.60 |
| Hospitalized | |||||
| 0.88 (0.74, 1.06) | 0.17 | 0.02 (–0.12, 0.16) | 0.79 | 0.93 (0.63, 1.36) | 0.70 |
| Extended hospitalized | |||||
| 1.0 (0.89, 1.12) | 0.93 | –0.01 (–0.10, 0.08) | 0.84 | 0.97 (0.75, 1.26) | 0.83 |
| Severe disease | |||||
| 0.79 (0.59, 1.05) | 0.11 | 0.06 (–0.15, 0.28) | 0.58 | 0.93 (0.50, 1.72) | 0.81 |
| Extended severe disease | |||||
| 0.99 (0.84, 1.16) | 0.87 | –0.01 (–0.14, 0.12) | 0.92 | 0.97 (0.68, 1.38) | 0.87 |
Odds ratios are presented for a decrease in 1 standard deviation in angiotensin-converting enzyme level. CI, confidence interval.
Using the PhenoScanner tool, we identified two SNPs that were associated with body mass index and pulmonary-function tests—traits that have been associated with infectious-disease outcomes in previous studies.36–38 Since it is possible that these SNPs may reflect horizontal pleiotropy effects, they were excluded in a sensitivity analysis. Doing so rendered similar results for susceptibility (OR: 1.02, 95% CI: 0.95, 1.11, P = 0.54), extended susceptibility (OR: 1.02, 95% CI: 0.95, 1.09, P = 0.62), hospitalization (OR: 0.88, 95% CI: 0.73, 1.06, P = 0.19), extended hospitalization (OR: 0.98, 95% CI: 0.87, 1.11, P = 0.78), severe disease (OR: 0.78, 95% CI: 0.58, 1.06, P = 0.12) and extended severe-disease phenotypes (OR: 0.99, 95% CI: 0.82, 1.18, P = 0.88) (Table 5).
Table 5.
MR results from sensitivity analysis with linkage-disequilibrium coefficient (r2) <10% and without SNPs at risk of pleiotropy
| Inverse-variance weighted MR meta-analysis |
MR-Egger meta-analysis |
||||
|---|---|---|---|---|---|
| Odds ratio (95% CI) | p-value | Intercept | Alpha p-value | Odds ratio (95% CI) | Odds ratio p-value |
| Susceptibility | |||||
| 1.02 (0.95, 1.11) | 0.54 | –0.02 (–0.08, 0.04) | 0.59 | 0.98 (0.84, 1.16) | 0.85 |
| Extended susceptibility | |||||
| 1.02 (0.95, 1.09) | 0.62 | 0.01 (–0.05, 0.06) | 0.77 | 1.04 (0.89, 1.21) | 0.64 |
| Hospitalized | |||||
| 0.878 (0.73, 1.06) | 0.19 | 0.02 (–0.12, 0.16) | 0.75 | 0.93 (0.63, 1.37) | 0.73 |
| Extended hospitalized | |||||
| 0.982 (0.87, 1.11) | 0.78 | –0.005 (–0.10, 0.09) | 0.93 | 0.97 (0.74, 1.27) | 0.84 |
| Severe disease | |||||
| 0.784 (0.58, 1.06) | 0.12 | 0.06 (–0.16, 0.28) | 0.58 | 0.92 (0.50, 1.70) | 0.80 |
| Extended severe disease | |||||
| 0.986 (0.82, 1.18) | 0.88 | –0.007 (–0.15, 0.14) | 0.93 | 0.97 (0.65, 1.44) | 0.88 |
Odds ratios are presented for a decrease in 1 standard deviation in angiotensin-converting enzyme level. CI, confidence interval.
The same conclusions were reached when using the cis-pQTL SNP (rs4344) from the AGES Reykjavik study22 (Table 6) and for the Host(a)ge cohort (Supplementary Table 2, available as Supplementary data at IJE online).
Table 6.
MR results from sensitivity analysis using a cis-pQTL from the Icelandic population22
| Inverse-variance weighted MR meta-analysis | |
|---|---|
| Odds ratio (95% CI) | p-value |
| Susceptibility | |
| 0.98 (0.89, 1.09) | 0.76 |
| Extended susceptibility | |
| 1.0 (0.93, 1.08) | 0.97 |
| Hospitalized | |
| 0.86 (0.66, 1.11) | 0.23 |
| Extended hospitalized | |
| 0.91 (0.79, 1.05) | 0.21 |
| Severe disease | |
| 0.75 (0.50, 1.14) | 0.18 |
| Extended severe disease | |
| 0.90 (0.71, 1.13) | 0.35 |
Note that the effect can only be used to infer the direction of effect, as a non-linear Yeo-Johnson transform was used on measured angiotensin-converting enzyme levels. CI, confidence interval.
Discussion
Using large populations and genetic variants with large effects on serum-ACE levels, we found that genetically decreased serum-ACE levels did not increase COVID-19 susceptibility or severity. Moreover, the narrow 95% CIs around the null observed in both the primary MR analyses and the multiple-sensitivity analyses suggest that, even if there were an underlying effect of decreased serum-ACE levels on these outcomes, the magnitude of this effect would not be clinically relevant. Lastly, since the same genetic variants that decrease serum-ACE levels were associated with a diagnosis of hypertension and use of ACE inhibitors, it is likely that these SNPs reflect a physiological effect of ACE. Taken together, our findings suggest that individuals should not stop these medications to prevent COVID-19 outcomes.
Multiple published traditional epidemiology studies have also not demonstrated harm with the use of ACE inhibitors.6–8,39 However, these studies were all retrospective and likely to be confounded by multiple unmeasured or improperly controlled for variables. Despite their large sample sizes, their confidence intervals were also wide, suggesting some uncertainty to these findings. By using naturally occurring randomization, we have greatly decreased the risk of bias due to confounding. Whereas randomized trials of ACE-inhibitor discontinuation are underway, patients at highest risk of severe disease are also likely to be those who could have adverse outcomes due to stopping ACE inhibitors (e.g. patients with heart failure), which might limit enrolment and bias the results in the direction of the null. For now, our study therefore provides evidence assessing the role of ACE inhibition during the pandemic.
Nonetheless, our study has multiple limitations. First, some of our analyses were likely underpowered to detect smaller COVID-19-outcome-effect sizes due to variations in serum-ACE levels. This is most pronounced in the severe-disease phenotypes, for which sample sizes were considerably smaller than those for the other phenotypes. Nevertheless, we can likely rule out large effect sizes from variations in ACE levels on COVID-19 severity. Given the known benefits from ACE inhibitors, our overall conclusions remain unchanged.
Second, we measured genetically determined variation in ACE levels and use this to infer the effect of ACE inhibitors on COVID-19 susceptibility and severity. For this to be relevant, we must assume that the patient’s ACE inhibitor has reached pharmacological steady state and that ACE levels have also reached their new lowered baseline. Therefore, our study cannot be used to make any recommendations on patients who recently started or stopped taking ACE inhibitors. However, since genetically decreased ACE levels are associated with better cardiovascular-disease outcomes and lower blood pressure,17,40 genetically lower ACE levels are likely a reasonable proxy for chronic use of ACE inhibitors. Hence, our results show that COVID-19 is unlikely to be a valid reason to stop ACE-inhibitor use.
Third, our primary analysis used cohorts of mixed genetic ancestry. Specifically, the ORIGIN trial cohort included patients of both Latin American and European ancestry, and it is possible that the effect of our instruments on either ACE levels or COVID-19 severity might differ between populations. However, allele frequencies were similar in both Latin American and European populations, and most were common, suggesting that extreme variations in the effect of ACE levels on the risk of severe infections are unlikely. Moreover, we also performed an analysis using a cis-pQTL from a strictly European-ancestry population (Iceland) and obtained similar results.
Finally, like all MR studies, our results may have been affected by unmeasured horizontal pleiotropy. To assess for this bias, we used MR-Egger analysis and used PhenoScanner to remove SNPs at the highest risk of pleiotropy. Most importantly, we only used cis-SNPs. Given their close distance to the ACE gene, these are less likely to act on the outcomes, independently of ACE levels. Hence, whereas the risk of a residual horizontal pleiotropic effect cannot be ruled out, we believe it is unlikely to change the conclusions of this study in a clinically meaningful way.
In conclusion, genetically lowered circulating ACE levels are not associated with COVID-19 susceptibility and severity. In balance, current evidence does not support the need to discontinue ACE inhibitors in order to reduce the risk of susceptibility and severity of COVID-19.
Supplementary data
Supplementary data are available at IJE online.
Author contributions
Conception and design: G.B.L., T.N., J.B.R. Data acquisition: A.R., S.A. Data analyses: G.B.L., T.N. Interpretation: G.B.L., T.N., V.M., A.R., S.A., S.Z., Y.C., V.F., J.B.R. Computational resources and support: V.F., J.B.R. Writing original draft: G.B.L., T.N. All authors were involved in reviewing the manuscript and critically reviewed its content. All authors gave final approval of the version to be published. G.B.L. and J.B.R. are the guarantors.
Funding
The Richards research group is supported by the Canadian Institutes of Health Research, the Lady Davis Institute of the Jewish General Hospital, the Canadian Foundation for Innovation, the NIH Foundation, Cancer Research UK and the Fonds de Recherche Québec Santé (FRQS). T.N. is supported by Research Fellowships of Japan Society for the Promotion of Science (JSPS) for Young Scientists and JSPS Overseas Challenge Program for Young Researchers. J.B.R. is supported by a FRQS Clinical Research Scholarship. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. These funding agencies had no role in the design, implementation or interpretation of this study. This research has been conducted using the UK Biobank resource (project number: 27449).
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank resource (project number: 27449). Members of the COVID-19 HGI (listed in Supplementary Table 5, available as Supplementary data at IJE online) and members of GEN-COVID Multicenter Study (listed in Supplementary Table 6, available as Supplementary data at IJE online) are acknowledged for their involvement in contributing to the publicly available COVID-19 HGI summary statistics round 3 (meta-analysis release date of 2 July 2020). The study was approved by individual research ethics boards from participating cohorts used in this study. Data are openly available through the COVID-19 Host Genome Initiative.
Conflict of interest
All authors have completed the ICMJE uniform disclosure form at http://www.icmje.org/coi_disclosure.pdf and declare that J.B.R. has served as an advisor to GlaxoSmithKline and Deerfield Capital for programmes unrelated to the research presented here. All other authors have nothing to disclose.
References
- 1. Hoffmann M, Kleine-Weber H, Schroeder S. et al. SARS-CoV-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turner AJ. Chapter 25—ACE2 cell biology, regulation, and physiological functions. In: Unger T, Steckelings UM, dos Santos RAS (eds). The Protective Arm of the Renin Angiotensin System (RAS). Boston: Academic Press, 2015, pp. 185–89. [Google Scholar]
- 3. Herman LLP, Annamaraju P, Bashir K.. Angiotensin Converting Enzyme Inhibitors (ACEI). Treasure Island, FL: StatPearls Publishing, 2020. [PubMed] [Google Scholar]
- 4. Hales CS, Martin CB, Kohen D.. Prescription Drug Use among Adults Aged 40–79 in the United States and Canada. Hyattsville, MD: National Center for Health Statistics, 2019. [PubMed] [Google Scholar]
- 5. Zheng Y-Y, Ma Y-T, Zhang J-Y, Xie X.. COVID-19 and the cardiovascular system. Nat Rev Cardiol 2020;17:259–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fosbøl EL, Butt JH, Østergaard L. et al. Association of angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use with covid-19 diagnosis and mortality. JAMA 2020;324:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mancia G, Rea F, Ludergnani M, Apolone G, Corrao G.. Renin-angiotensin-aldosterone system blockers and the risk of covid-19. N Engl J Med 2020;382:2431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reynolds HR, Adhikari S, Pulgarin C. et al. Renin-angiotensin-aldosterone system inhibitors and risk of covid-19. N Engl J Med 2020;382:2441–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sommerstein R, Kochen MM, Messerli FH, Gräni C.. Coronavirus disease 2019 (COVID-19): do angiotensin-converting enzyme inhibitors/angiotensin receptor blockers have a biphasic effect? J Am Heart Assoc 2020;9:e016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Statement from the American Heart Association, the Heart Failure Society of America and the American College of Cardiology. Patients Taking Ace-i and Arbs Who Contract COVID-19 Should Continue Treatment, Unless Otherwise Advised by Their Physician. https://newsroom.heart.org/news/patients-taking-ace-i-and-arbs-who-contract-covid-19-should-continue-treatment-unless-otherwise-advised-by-their-physician (23 June 2020, date last accessed).
- 11.International Society of Hypertension. A Statement from the International Society of Hypertension on COVID-19. https://ish-world.com/news/a/A-statement-from-the-International-Society-of-Hypertension-on-COVID-19 (23 June 2020, date last accessed).
- 12. Zhang P, Zhu L, Cai J. et al. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin ii receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ Res 2020;126:1671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J, Wang X, Chen J, Zhang H, Deng A.. Association of renin-angiotensin system inhibitors with severity or risk of death in patients with hypertension hospitalized for coronavirus disease 2019 (COVID-19) infection in Wuhan, China. JAMA Cardiol 2020;5:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith GD, Ebrahim S.. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 15. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 16. Hemani G, Bowden J, Davey Smith G.. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 2018;27:R195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Donnell CJ, Lindpaintner K, Larson MG. et al. Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in the Framingham Heart Study. Circulation 1998;97:1766–72. [DOI] [PubMed] [Google Scholar]
- 18. Ljungberg L, Alehagen U, Länne T. et al. The association between circulating angiotensin-converting enzyme and cardiovascular risk in the elderly: a cross-sectional study. J Renin Angiotensin Aldosterone Syst 2011;12:281–89. [DOI] [PubMed] [Google Scholar]
- 19. Davey Smith G, Hemani G.. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pigeyre M, Sjaarda J, Chong M. et al. ACE and type 2 diabetes risk: a Mendelian randomization study. Diabetes Care 2020;43:835–42. [DOI] [PubMed] [Google Scholar]
- 21. Gerstein HC, Bosch J, Dagenais GR. et al. Basal insulin and cardiovascular and other outcomes in dysglycemia. N Engl J Med 2012;367:319–28. [DOI] [PubMed] [Google Scholar]
- 22. Emilsson V, Ilkov M, Lamb JR. et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361:769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Auton A, Brooks LD, Durbin RM. et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Machiela MJ, Chanock SJ.. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (Oxford, England )2015;31:3555–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wuttke M, Li Y, Li M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 2019;51:957–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah S, Henry A, Roselli C. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warren HR, Evangelou E, Cabrera CP. et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet 2017;49:403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y, Byrne EM, Zheng Z. et al. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun 2019;10:1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamat MA, Blackshaw JA, Young R. et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics (Oxford, England )2019;35:4851–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staley JR, Blackshaw J, Kamat MA. et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics (Oxford, England) 2016;32:3207–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The COVID-19 Host Genetics Initiative. A global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet 2020;28:715–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willer CJ, Li Y, Abecasis GR.. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England )2010;26:2190–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ellinghaus D, Degenhardt F, Bujanda L. et al. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med 2020;383:1522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walker VM, Davies NM, Hemani G. et al. Using the MR-base platform to investigate risk factors and drug targets for thousands of phenotypes. Wellcome Open Res 2019;4:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Butler-Laporte G, Harroud A, Forgetta V, Richards JB, Elevated body-mass index is associated with an increased risk of infectious disease admissions and mortality: a Mendelian randomization study. Clin Microbiol Infect 2020, Jun 24:S1198-743X(20)30356-6. doi: 10.1016/j.cmi.2020.06.014. [Epub ahead of print.]. [DOI] [PubMed] [Google Scholar]
- 37. Winter-Jensen M, Afzal S, Jess T, Nordestgaard BG, Allin KH.. Body mass index and risk of infections: a Mendelian randomization study of 101,447 individuals. Eur J Epidemiol 2020;35:347–54. [DOI] [PubMed] [Google Scholar]
- 38. Sabia S, Shipley M, Elbaz A. et al. Why does lung function predict mortality? Results from the Whitehall II Cohort Study. Am J Epidemiol 2010;172:1415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raisi-Estabragh Z, McCracken C, Ardissino M. et al. Renin-angiotensin-aldosterone system blockers are not associated with coronavirus disease 2019 (COVID-19) hospitalization: study of 1,439 UK Biobank cases. Front Cardiovasc Med 2020;7:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cambien F, Poirier O, Lecerf L. et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature 1992;359:641–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.