Abstract
The circadian clock exerts temporal coordination of metabolic pathways. Clock disruption is intimately linked with the development of obesity and insulin resistance, and our previous studies found that the essential clock transcription activator, Brain and Muscle Arnt-like 1 (Bmal1), is a key regulator of adipogenesis. However, the metabolic consequences of chronic shiftwork on adipose tissues have not been clearly defined. Here, using an environmental lighting-induced clock disruption that mimics rotating shiftwork schedule, we show that chronic clock dysregulation for 6 months in mice resulted in striking adipocyte hypertrophy with adipose tissue inflammation and fibrosis. Both visceral and subcutaneous depots display enlarged adipocyte with prominent crown-like structures indicative of macrophage infiltration together with evidence of extracellular matrix remodeling. Global transcriptomic analyses of these fat depots revealed that shiftwork resulted in up-regulations of inflammatory, adipogenic and angiogenic pathways with disruption of normal time-of-the-day-dependent regulation. These changes in adipose tissues are associated with impaired insulin signaling in mice subjected to shiftwork, together with suppression of the mTOR signaling pathway. Taken together, our study identified the significant adipose depot dysfunctions induced by chronic shiftwork regimen that may underlie the link between circadian misalignment and insulin resistance.
Keywords: circadian clock, adipose tissue, inflammation, shiftwork, insulin resistance
1. INTRODUCTION
The circadian clock, an evolutionarily-conserved transcriptional-translational network that generates the ~24 hour rhythms, is intimately linked with metabolic regulation (Schibler and Sassone-Corsi 2002, Dibner, Schibler et al. 2010). Circadian clock controls key steps of metabolic pathways to ensure coordination of nutrient metabolism with environmental cues (Panda, Antoch et al. 2002, Bass and Takahashi 2010). Accumulating evidence from experimental models and epidemiological studies indicates that disruption of clock regulation leads to the development of obesity and insulin resistance (Parkes 2002, Turek, Joshu et al. 2005, Scheer, Hilton et al. 2009, Karatsoreos, Bhagat et al. 2011, Pan, Schernhammer et al. 2011). However, the underlying mechanisms linking circadian disruption and metabolic diseases remains poorly understood.
The circadian clock network in our body is a hierarchical system that consists of the central clock residing in the suprachiasmatic nuclei (SCN) and peripheral clocks present in nearly all tissue and cell types (Takahashi 2017). Under normal 24-hour light-dark cycles, peripheral clock circuits are driven by the SCN clock that responds to light signals transmitted through the retino-hypothalamic tract. However, peripheral clock rhythms, particularly metabolic tissue clock systems, can be uncoupled from the central clock by feeding-associated signals (Bass and Takahashi 2010). Through the integration of central clock-derived metabolic or humoral signals, coordination between central clock and peripheral clocks-controlled metabolic processes is critical to maintain whole-body metabolic homeostasis. In our modern lifestyle, frequent misalignment of sleep/activity cycles with endogenous clock cycles creates wide-spread circadian dys-synchrony (Roenneberg, Allebrandt et al. 2012), which may contribute to the current epidemics of metabolic diseases. Despite the current strong evidence supporting the link between circadian misalignment and metabolic abnormalities, particularly obesity and insulin resistance, a causal relationship has not been formally tested in animal models that recapitulates the long-term consequences of clock disruption (Pan, Schernhammer et al. 2011, Buxton, Cain et al. 2012).
Large-scale epidemiological studies indicate that shiftwork exposure is associated with diabetes risk and indexes of obesity (van Amelsvoort, Schouten et al. 1999, Pan, Schernhammer et al. 2011, Vetter, Devore et al. 2015). In addition, imposing circadian misalignment in controlled lighting environment revealed that acute alteration of sleep-activity periods leads to impaired insulin sensitivity and reduced energy expenditure (McHill, Melanson et al. 2014). Nonetheless, studies to date are mostly limited to the acute metabolic consequences of circadian shifts, while its long-term adverse effects, as demonstrated by large-scale epidemiology studies, remains to be addressed. In addition, potential distinct contributions of visceral and subcutaneous fat depots and underlying mechanisms involved in impaired energy balance under circadian misalignment have not been carefully dissected. As a major site for energy storage, mobilization and endocrine regulation, how adipose tissue adapts to shift regimens has not been defined.
Adipose tissue are energy storage organs that are key determinant of whole-body metabolic homeostasis, and functional peripheral clocks are present in various adipose depots (Zvonic, Ptitsyn et al. 2006, Wu, Zvonic et al. 2007, Otway, Frost et al. 2009). Many clock components, including the essential clock activator Brain and Muscle Arnt-like 1 (Bmal1), and the key clock repressor Rev-erbα, directly participate in adipogenesis (Fontaine, Dubois et al. 2003, Grimaldi, Bellet et al. 2010, Guo, Chatterjee et al. 2012). These and other core clock regulators can modulate brown adipocyte development that consequently impacts brown fat thermogenic capacities (Nam, Chatterjee et al. 2015, Nam, Guo et al. 2015).
In the current study, we applied an environmentally-induced clock disruption mimicking shiftwork for 6 months to uncover its metabolic consequences specifically in adipose tissue. Through global gene expression profiling together with functional analysis, we show that chronic clock dysregulation results in striking adipose tissue hypertrophy with overt inflammatory response and fibrotic sequala that predispose to insulin resistance.
2. MATERIAL AND METHODS
2.1. Animals
Male C57/BL6 mice were purchased from the Jackson Laboratory and maintained on regular chow diet in the Rensselaer Polytechnic Institute vivarium rooms with dedicated lighting controls as specified. All experiments were approved by the IACUC committee of Rensselaer Polytechnic Institute (RPI). Mice were assigned randomly to the normal control or a rotating shift schedule defined as the shiftwork group. Mice in the normal control group were maintained in a regular lighting condition of 12-h light, 12-h dark cycle (lights on 5AM; lights off 5PM). The time of lights on was defined as Zeitgeber time zero (ZT0). The mice in the shiftwork group were subjected to a weekly rotating light schedule every week, with normal L/D cycle of lights on 5AM and lights off at 5PM from Monday to Thursday and reversed L/D cycle on Friday to Sunday of lights on at 5PM and lights off at 5AM.
2.2. Hematoxylin and eosin histology
Adipose tissues were collected and fixed with 10% neutral-buffered formalin for 72 hours, washed in 70% alcohol prior to embedding in paraffin. 10μm paraffin-embedded sections were processed for deparaffinization using xylene, rehydrated in ethanol, and stained with hematoxylin and eosin for histological analysis. Adipocyte size was calculated by outlining the adipocytes to measure area, and the average of the sum of three representative 10X fields from each mice of group were plotted for size distribution curve.
2.3. Immunofluorescence staining
Paraffin-embeded slides were de-paraffinized and processed for antigen retrieval by steaming the slides for 10 minutes. The slides were permeabilized by 0.2% Triton X-100 in PBS and blocked in 3% horse serum. Primary monoclonal F4/80 antibody (MA1-91124, Thermofisher, 1:100) was incubated at room temperature for 30 minutes with blocking solution. Images were captured using Echo Microscope at indicated magnifications.
2.4. Masson’s Trichrome
This staining was performed to identify collagen in adipose tissue fibrosis using parafin-embeded slides following de-parafinization. Trichrome staining were performed using the Masson's Trichrome Stain Kit according to manufacturer’s instructions (Polyscience Inc.). Briefly, slides were dehydrated in ethanol, and sequentially processed in Bouin’s fixative at 60°C for 1 hour, Weigert’s Iron hematoxylin for 10 minutes, Biebrich Scarlet-Acid Fuchsin for 5 minutes, Phosphotungstic/phosphomolybdic acid for 10 minutes, followed by washing in acetic acid.
2.5. Immunoblot analysis
Protein samples were extracted using RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor (Roche). 20-40 μg of total protein was resolved on SDS-PAGE gels followed by immunoblotting after nitrocellulose membrane transfer. Primary and HRP-conjugated secondary antibodies were applied and developed by enhanced chemiluminescence substrate (Supersignal; Thermo Scientific). The primary antibodies and dilutions used were: C/EBPβ 1:1,000 (Santa Cruz, sc-150), PPARγ 1:1,000 (Santa Cruz, sc-61): Akt 1:1,000 (Cell Signaling, 4691S), Phospho-Akt (Ser473) 1:1,000 (Cell Signaling, 4060S), Hsp90 1:2,000 (Cell signaling, 4874).
2.6. RNA-seq sequencing library preparation and sequencing with Illumina Hiseq2500
Total RNA was extracted using Trizol (Invitrogen) followed by Qiagen RNeasy miniprep column purification (Qiagen). Sequencing libraries were prepared with Kapa RNA mRNA HyperPrep kit (Kapa Biosystems) according to the manufacturer's protocol. Briefly, 250 ng of total RNA from each sample was used for polyA RNA enrichment. The enriched mRNA underwent fragmentation and first strand cDNA synthesis. The combined 2nd cDNA synthesis with dUTP and A-tailing reaction generated the resulting ds cDNA with dAMP to the 3’ ends. The barcoded adaptors were ligated to the double-stranded cDNA fragments. A 12-cycle of PCR was performed to produce the final sequencing library. The libraries were validated with the Agilent Bioanalyzer DNA High Sensitivity Kit and quantified with Qubit. RNA-seq libraries were sequenced on Illumina HiSeq 2500 with SR V4 Kit with the single read mode of 51cycle of read1 and 7 cycles of index read. Real-time analysis (RTA) 2.2.38 software was used to process the image analysis and base calling.
2.7. Bioinformatics Analysis of RNA-Seq data
RNA-seq sequencing reads were aligned with STAR software to mouse reference genome mm10 (Dobin, Davis et al. 2013). Unique read counts were quantified using HTSeq-count (Anders, Pyl et al. 2015), with GENCODE gene annotations. The RNA-seq read counts for genes were normalized and log-transformed into using limma, edgeR packages. P-values were calculated from raw counts using DESeq2 (Love, Huber et al. 2014), which also calculated the False Discovery Rate (FDR). Fold-change > 1.5, 50% FPKM > 0.1, unadjusted p-value < 0.05 and FDR < 0.25 were used as cut-off for differentially expressed genes (DEG). We performed Principle Component Analysis (PCA) for testing biological reproducibility within biological replicates. Global analysis heatmap were produced using heatmap.3 (heatmap.3.R) the gplots package (gplots, RColorBrewer) in R, using log2(FPKM + 0.1) values. Pearson Dissimilarity (1-Pearson correlation coefficient) was used as the distance metric for hierarchical clustering of rows and columns. Expression was centered so each gene had a mean of 0, and centered expression was capped at −3 and 3, for clustering and visualization. All RNA-seq datasets were deposited in NCBI GEO158996.
2.8. Functional annotation of enriched pathways
Gene Ontology (GO, (Ashburner, Ball et al. 2000)) and Kyoto Encyclopedia of Genes and Genomes (KEGG, (Kanehisa and Goto 2000)) pathway enrichment were calculated using DAVID (Dennis, Sherman et al. 2003), using gene symbols for differentially expressed genes. Additional systems-level analysis was performed in Ingenuity Pathway Analysis (www.ingenuity.com). For IPA analysis, we utilized the relaxed cutoff (Fold-change > 1.5, p-value < 0.05 and FDR<0.25) and the entire set of input genes served as the background for the enrichment tests. This cutoff was selected to yield sufficient number of candidate DEGs in the enriched gene sets (Dennis, Sherman et al. 2003). For pathway enrichment analysis, p-value < 0.05 was considered significant, and positive or negative z-score was calculated by up-regulation or down-regulation DEGs.
2.9. Statistical analysis
Values were expressed as mean ± SEM with minimum of three biological replicates. For differential gene expression analysis, negative binomial test with a cutoff of the false discovery rate (FDR) <0.05 was used. For other analyses, two-way ANOVA test by Prism software version 8 (GraphPad) was used and a p-value < 0.05 was considered significant.
3. RESULTS
3.1. Environmental clock disruption induced by a shiftwork regimen promotes adipocyte hypertrophy in visceral and subcutaneous fat depots
Based on our previous work in genetic models demonstrating that adipogenesis is under the control of circadian clock regulators (Guo, Chatterjee et al. 2012, Nam, Chatterjee et al. 2015, Nam, Guo et al. 2015), in the current study, we tested whether environmental lighting shift-induced clock disruption mimicking shiftwork affects adipose tissue growth. We designed a rotating shiftwork schedule with reversed L/D cycles for three days (lights on- 5PM, off-5 AM) every week following four days of normal L/D cycles (lights on-5 AM, off-5 PM), as shown in Fig. 1A. This shiftwork regimen is designed to elicit central and peripheral circadian clock dys-synchrony that resembles environmental circadian misalignment, and we previously demonstrated that a similar 5-week regimen caused β-cell clock dysregulation (Lee, Moulik et al. 2013). For analysis with normal control under a regular L/D schedule at comparable zeitgeber time (ZT) of the day, samples were collected on the second day of the normal L/D cycle in both groups at 8AM (ZT3) and 5PM (ZT12) as indicated.
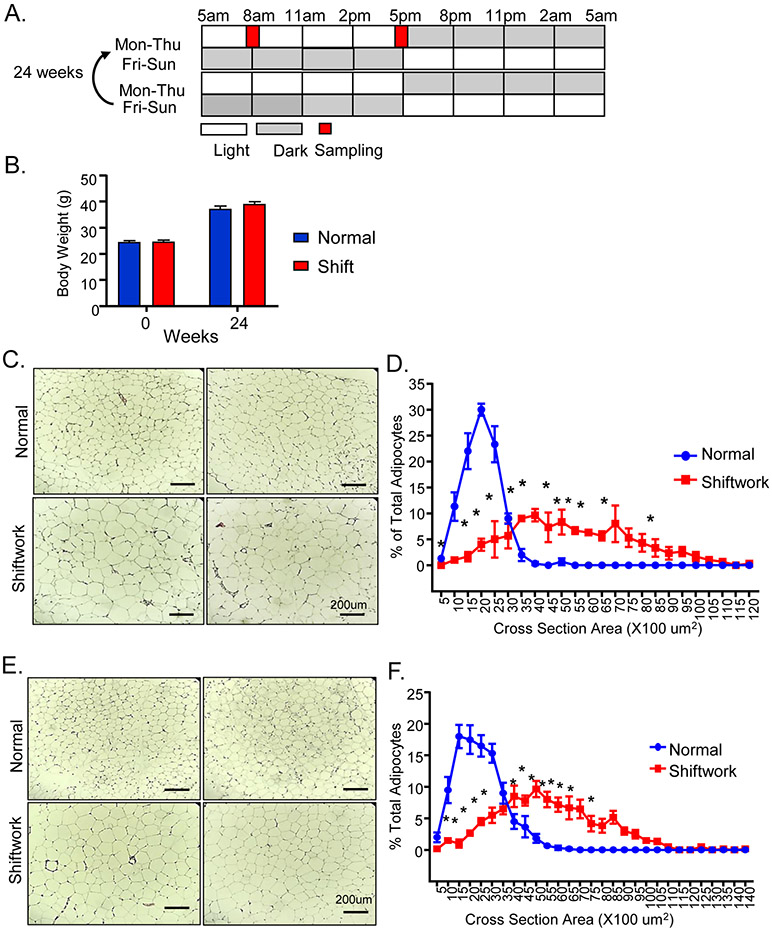
Figure 1.
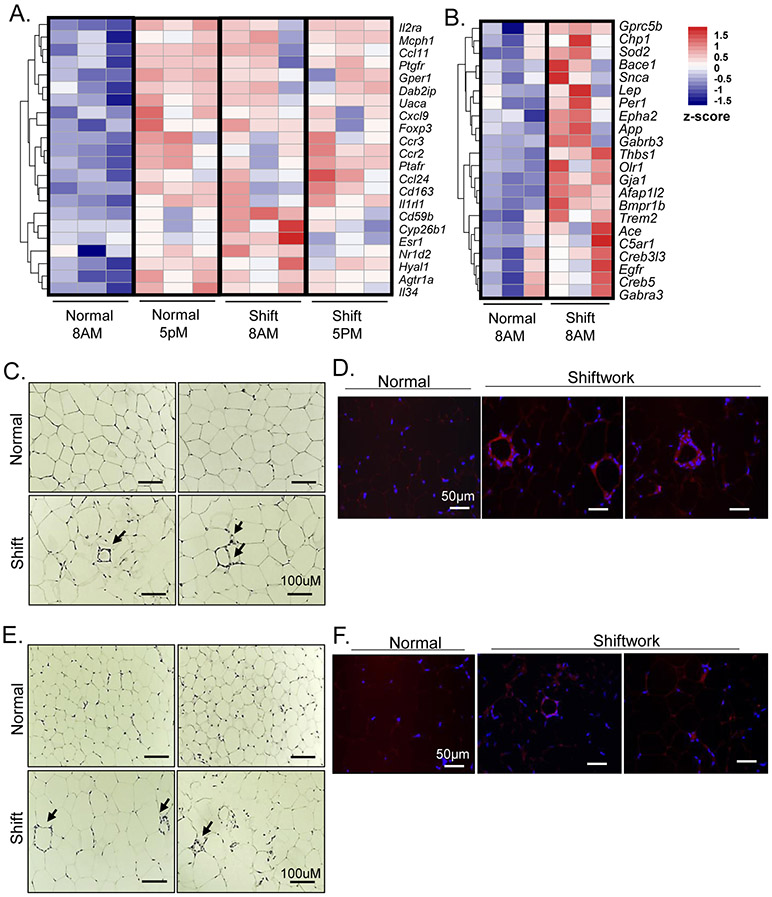
Chronic rotating shiftwork schedule for 6 months promotes adipocyte hypertrophy in adipose depots. (A) Schematic representation of weekly rotating lighting shift regimen in the shiftwork group with normal L/D from Monday to Thursday and reversed L/D cycle from Friday to Sunday. Two consecutive weeks were shown and the weekly rotation was maintained for 24 weeks. Samples were collected at indicated 8AM and 5PM on Tuesday with normal L/D lighting for both normal and shiftwork groups. (B) Body weight of normal control and shiftwork group mice at beginning of shift regimen and after 6 months (n=12/group). (C) Representative H/E histology after 6 months of shiftwork, and (D) histogram of adipocyte size distribution of eWAT (n=6/group). (E) Representative H/E histology and (F) adipocyte size distribution of iWAT (n=6/group). For adipocyte size measurement, three 10X fields were counted in each sample of normal or shiftwork groups (n=6). *: P≤0.05 or **: P≤0.01 by ANOVA.
We first determined the body weight gain over the six months subjected to the shiftwork regimen. Both control and the shiftwork group mice gained over more than 50% body weight over this period, and there were no differences in body weight between groups (Fig. 1B). Surprisingly, histological analysis of the visceral fat depot, the epididymal white adipose tissue (eWAT), revealed significantly enlarged adipocytes in mice in the shiftwork group (Fig. 1C). Quantification of eWAT adipocyte size distribution indicated a marked shift to larger adipocytes (Fig. 1D). Adipocyte hypertrophy was similarly observed in subcutaneous fat depot, the inguinal white adipose tissue (iWAT, Fig. 1E), with substantially skewed size distribution toward larger adipocytes (Fig. 1F). In addition, we noticed typical crown-like structures comprised of macrophages surrounding apoptotic adipocytes that are abundant in both the eWAT and iWAT in the shiftwork group, while they are rarely present in the controls.
3.2. Global transcriptomic profiling identified time-of-day differential gene expression in eWAT and disruption by shiftwork
To determine the mechanism underlying adipocyte hypertrophy induced by chronic shiftwork, we performed global transcriptomic profiling by RNA-Seq analysis of visceral and subcutaneous adipose depots. In eWAT, we also determined time-of-day-dependent differential gene expressions at ZT3 (8AM) and ZT12 (5PM) in control and the shiftwork groups (Fig. 2A). Interestingly, among all comparisons we found the highest number of significantly up- and down-regulated genes between ZT3 vs. ZT12 timepoints in normal controls under the 12L/12D condition than other comparisons, suggesting a strong time-of-the-day dependence on gene regulation in visceral fat depot. These differentially expressed genes (DEGs) were shown in Volcano plot in Fig. 2B. Notably, in the shiftwork group, we also observed significant differential gene expressions between ZT3 and ZT12 time points (Fig. 2A). Intriguingly, when we compared control and shiftwork groups, we found that gene expression alterations induced by shiftwork were most evident at ZT3 but markedly dampened at ZT12, with 1,589 genes differentially expressed at ZT3 as compared to only 176 genes at ZT12 (Fig. 2A). Using Ingenuity Pathway Analysis (IPA), we identified the top 15 differentially expressed pathways between ZT3 and ZT12 in the control group, and this analysis identified that inflammatory response and cell-cell communications are major enriched processes that display significant time-dependent regulation in eWAT (Fig. 2C). Various processes involved in inflammation, including inflammatory signaling, leucocyte extravasation, complement system macrophage phagocytosis and eNOS signaling, were up-regulated at ZT12 (Fig. 2C). Additional components related to immune regulation, including antigen presentation, B cell development, T cell and B cell signaling and Th1 and Th2 activation pathways, were over-represented in this comparison, although for these genes there were no evident patterns of activation or inhibition. Additional processes involved in cell-cell interactions, including Wnt/β-catenin and actin cytoskeleton signaling, many are known genes under circadian control as we identified previously (Guo, Chatterjee et al. 2012, Liu, Xiong et al. 2020), were elevated at ZT12. In contrast, key processes in cell-extracellular communications, including paxillin and integrin signaling, were suppressed at this time point.
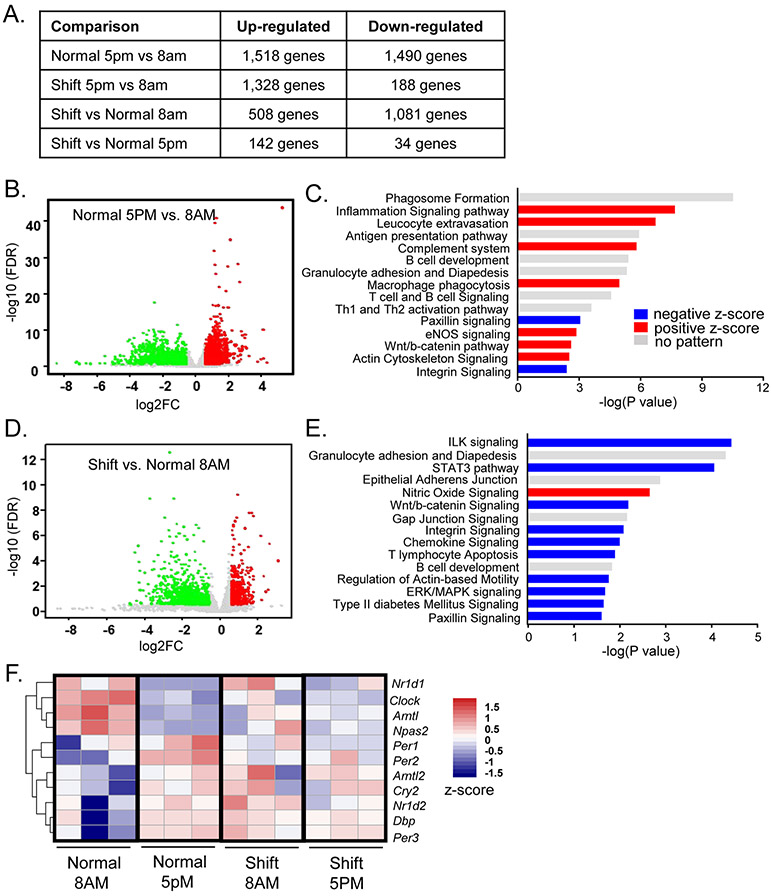
Figure 2.
RNA-seq analysis of chronic shiftwork-induced differentially expressed pathways in eWAT. (A) Hierarchical clustering of differentially expressed genes identified by RNA-seq of the four groups of eWAT samples analyzed, 8AM shiftwork, 8AM normal control, 5PM shiftwork, 5PM normal control (n=3/group). Each column represents a sample and each row corresponds to the annotate transcript with expression level shown according to the color scale. (B, C) Volcano plot of time-of-the-day dependent differentially expressed genes (B), and Ingenuity Pathway Analysis of top 15 differentially-regulated pathways (C) in eWAT between 5PM and 8AM in normal control groups (n=3/group). (D, E) Volcano plot of differentially expressed genes (D), and top 15 differentially-regulated canonical pathways induced by shiftwork as compared to normal controls in eWAT at 8AM (n=3/group) as identified by Ingenuity Pathway Analysis (−2.0 <Z score< 2.0) (E). Red horizontal bars indicate positive and blue for negative Z-score. (F) Heatmap representation of circadian clock gene regulation by shiftwork regimen in eWAT (8AM Shift/Normal). (E, F) Volcano plot of differentially expressed genes induced by shift vs. controls at 5PM (E, n=3/group) and time-of-the-day dependent differentially expressed genes in shiftwork group at 5PM as compared to 8AM (F, n=3/group). (G) Heatmap presentation of differentially regulated circadian clock genes by shiftwork regimen in eWAT (8AM Shift/Normal). Expression levels are shown according to color scale.
We next analyzed pathways affected by shiftwork at ZT3, a time when most significant gene regulation was detected as compared to ZT12. The overall differential gene regulation between the shiftwork and the control group was presented by Volcano plot in Fig. 2D. Analysis of top pathways altered by chronic shift revealed a striking overlap with processes with time-of-the-day regulation, including inflammation-related pathways such as granulocyte adhesion and diapedesis, T lymphocyte apoptosis, chemokine signaling and B cell development (Fig. 2E). These results reveal that global disruption of normal circadian gene expression in the shiftwork cohort persists despite acute normalization of lighting at the time of sample collection. Shiftwork suppressed cell-extracellular matrix interactions, including integrin-linked kinase (Ilk), Wnt/β-catenin, integrin, paxillin signaling, and regulation of actin-based motility that is downstream to integrin signaling pathway were observed. Notably, Type II diabetes-related processes in eWAT were down-regulated in mice subjected to shift regimen. As expected, specific time-of-the-day regulations of core circadian clock and direct clock-controlled genes (CCGs) were evident between 8AM and 5PM (Fig. 2F). However, this circadian pattern of clock gene expression in normal control was clearly abolished in the shiftwork group, even when sampled during normal L/D cycles with identical light entrainment. This finding implicates chronic rotating shift disruption of peripheral molecular clock oscillations that are not reversed by acutely reestablished external lighting.
3.3. Differential gene expression induced by shiftwork in subcutaneous adipose tissue
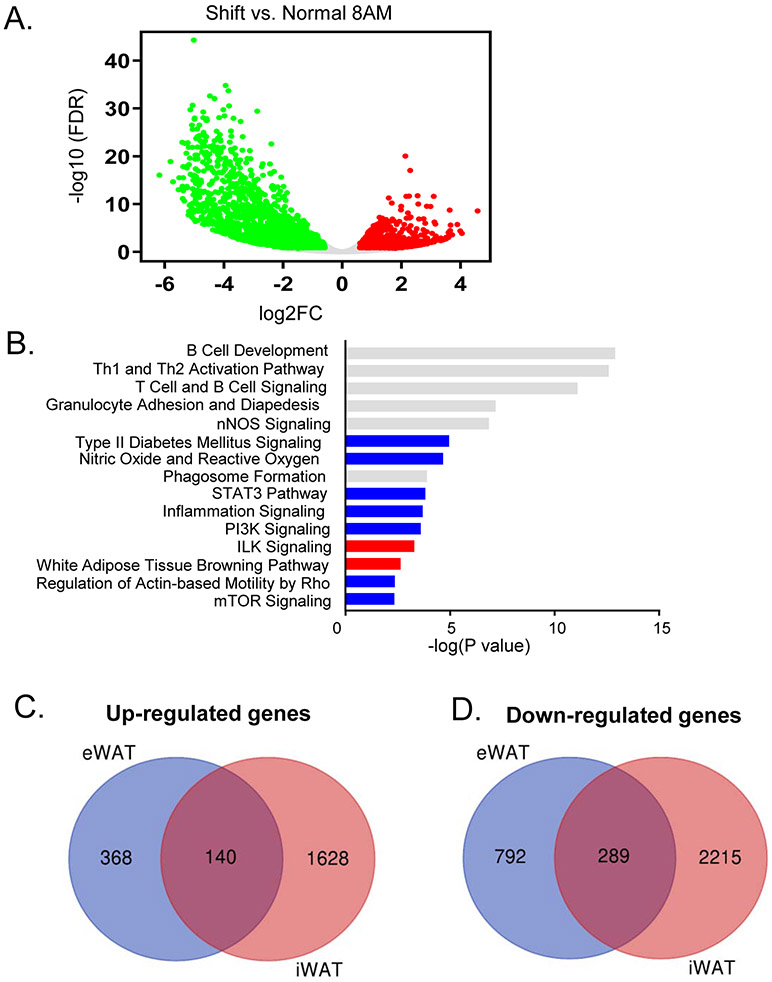
Using the inguinal subcutaneous adipose tissue as a representative beige fat depot, we determined shiftwork-modulated pathways and the potential overlap with differentially regulated processes identified in visceral fat. Differentially regulated genes by shiftwork in iWAT at ZT3 were plotted in Fig. 3A. IPA analysis of top shift-modulated pathways in iWAT indeed reveal significant overlap with eWAT (Fig. 3B), particularly processes involved in inflammation and immune regulation. Similar to eWAT, Type II diabetes and mTOR signaling pathways were down-regulated by shiftwork, suggesting insulin resistance in this tissue. Global comparison of up- and down-regulated genes in iWAT and eWAT induced by shift regimen revealed the extent of the overlap, as demonstrated in Fig. 3C & 3D, although significant more abundant differentially-regulated transcripts were detected in iWAT. Interestingly, among either overlapping or non-overlapping genes between iWAT and eWAT, the enriched pathways detected were comparable, with immune functions and inflammatory response identified as the top enriched processes in both fat depots.
Figure 3.
RNA-seq analysis of chronic shiftwork-induced differentially expressed pathways in subcutaneous beige fat. (A, B) Volcano plot of differentially expressed genes induced by shiftwork (A), and top 15 differentially-regulated pathways in 8AM iWAT samples as identified by IPA (B) between shiftwork and normal control groups (n=3/group). (C, D) Venn diagram depicting the comparison of up-regulated (C), and down-regulated genes (D) between iWAT and eWAT 8AM samples (n=3/group).
3.4. Time-of-day regulation of adipocyte differentiation pathway and induction by chronic shiftwork
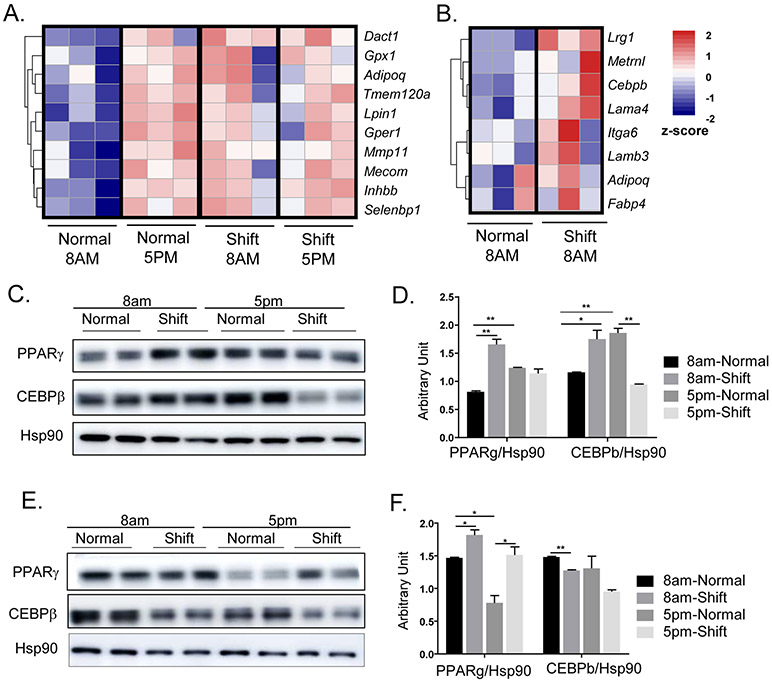
Chronic shiftwork induced marked adipocyte hypertrophy in visceral and subcutaneous fat depots. Adipose expansion during development of obesity involves both hypertrophy and addition of new adipocytes through adipogenesis. We found that one of the significantly up-regulated pathways enriched in the shiftwork group belongs to fat cell differentiation in eWAT, which included several genes involved in lipid metabolism (Fig. 4A). Interestingly, this pathway was found to display pronounced circadian regulation as indicated by consistently lower expression at ZT3 than at ZT12, suggesting increased adipogenic drive at onset of active cycle that coincides with feeding. Furthermore, this time-of-day-dependent regulation was abolished in the shiftwork group, with strikingly near uniform up-regulations at ZT3 that reached comparable levels observed in controls at ZT12. This persistently elevated levels of adipogenic drive induced by chronic shift may underlie, at least in part, adipose tissue expansion. In the iWAT, shift regimen resulted in a similarly up-regulated fat cell differentiation pathway suggesting augmented adipogenic response (Fig. 4B). We next validated the transcriptomic changes in eWAT and iWAT through examination of protein expressions of key adipogenic factors. Notably, PPARγ and C/EBPβ protein in eWAT displayed strong diurnal expression patterns that are higher at ZT12 than ZT3, consistent with the time-of-day fluctuations of their respective transcript levels (Fig. 4C & 4D). In mice undergoing shiftwork regimen, both PPARγ and C/EBPβ were markedly induced as compared to controls at ZT3, whereas this trend was reversed at ZT12 with suppressed expressions. Thus, the normal diurnal patterns of expression of both adipogenic factors was disrupted by shiftwork in eWAT. In contrast, PPARγ protein in iWAT was also induced in the shifted group at both at ZT3 and ZT12, although its diurnal pattern was reversed as compared to eWAT (Fig. 4E & 4F). C/EBPβ in iWAT did not exhibit circadian-time dependence, and was moderately inhibited by shiftwork. These findings collectively reveal the rhythmic expression of adipogenic pathway transcription regulators and a tendency toward augmented adipogenic response by chronic shift, a mechanism that may contribute to adipose tissue expansion.
Figure 4.
Chronic shiftwork induces augmented adipogenic response. (A, B) Heatmap presentation of differentially-expressed genes involved in fat cell differentiation pathway as identified by IPA analysis in 6-month shiftwork and normal control groups in eWAT (A), and (B) iWAT, (n=3/group). Transcripts are annotated and expression levels are shown according to color scale. (C, D) Immunoblot analysis (C), and quantification (D) of key adipogenic factors of eWAT in normal and shiftwork group at 8AM and 5PM (n=6/group, 3 pooled samples each lane). (E, F) Immunoblot analysis (E) and quantification (F) of key adipogenic factors of iWAT in normal and shiftwork 8AM samples (n=6/group, 3 pooled samples each lane). *: P≤0.05 or **: P≤0.01 by Student’s t test.
3.5. Chronic shiftwork induces inflammatory and fibrotic response in visceral and subcutaneous fat depots
Initial histological analysis revealed numerous crown-like structures present in fat depots of shiftwork cohort indicative of macrophage infiltration, characteristic of adipose tissue inflammation. Consistent with this observation, RNA-seq analysis identified significantly augmented expression of genes involved in inflammatory response enriched in both eWAT and iWAT. In visceral fat, this pathway displayed normally low expressions at ZT3 and was elevated at ZT12, exhibiting a diurnal expression profile (Fig. 5A). Chronic shift led to significantly higher expression of this pathway at ZT3 that largely resembles that of ZT12 with a loss of the diurnal pattern of expression. This pathway was also induced in iWAT of the shiftwork group (Fig. 5B). Detailed histological examination of eWAT in mice subjected to shift demonstrated the abundant crown-like structure characteristics of macrophage accumulation, in contrast to minimal crown-like structures detected in controls (Fig. 5C). Using a macrophage-specific surface marker F4/80 immunofluorescence, we identified macrophages surrounding apoptotic adipocytes that formed the crown-like structures that are easily found in eWAT of chronic shiftwork group, with very limited staining detected in the controls (Fig. 5D). Interestingly, iWAT of shiftwork group also displayed prominent crown structures (Fig. 5E) that can be verified by F4/80 immunostaining (Fig. 5F), but are barely identifiable in the control cohort. Thus, shiftwork elicited pronounced inflammatory response exemplified by abundant macrophage infiltration in visceral and subcutaneous adipose depots accompanying adipocyte hypertrophy.
Figure 5.
Chronic shiftwork induces inflammatory response. (A, B) Heatmap presentation of differentially-expressed genes involved in inflammatory pathway identified in 6-month shiftwork and normal control groups at 8AM and 5PM in eWAT (A), and (B) 8AM samples in iWAT (n=3/group). Transcripts are annotated and expression levels shown according to color scale. (C, D) Representative images of H/E staining (C), and F4/80 immunofluorescence staining of macrophages (D) with arrows indicating crown-like structure in eWAT in chronic shiftwork and normal controls. Scale bar: 100 μM. (E, F) Representative images of H/E staining (E), and F4/80 immunofluorescence staining of macrophages (F) with arrowhead indicating crown-like structures in iWAT in chronic shiftwork and normal control group. Scale bar: 50 μM.
3.6. Chronic shiftwork induces angiogenic and fibrotic response in visceral and subcutaneous fat depots
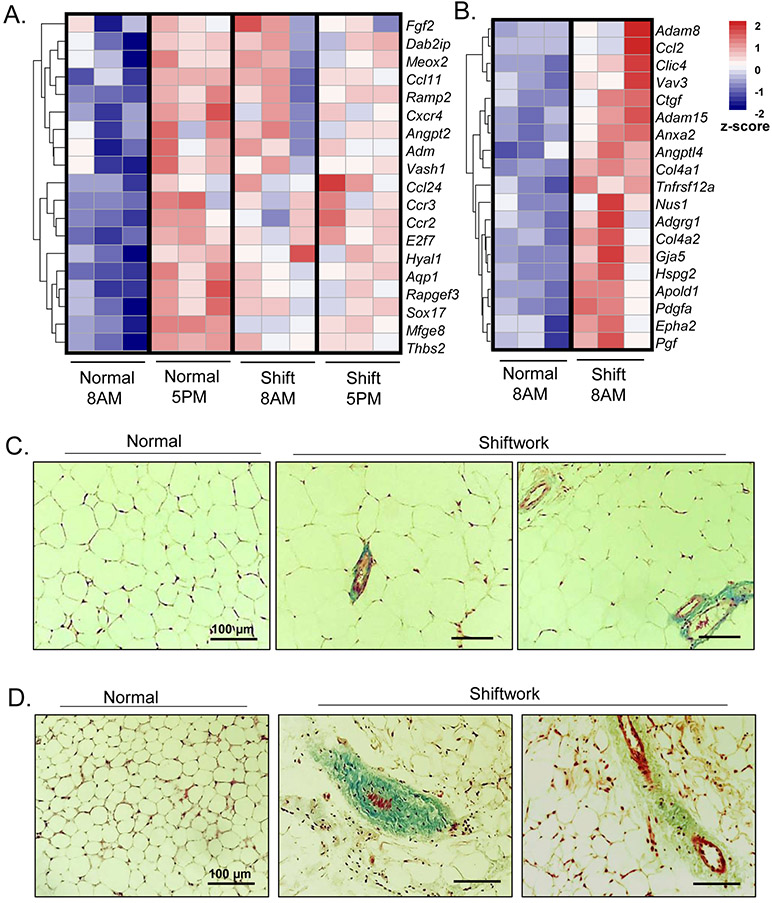
Macrophage infiltration and inflammatory milieu in adipose tissue ultimately leads to extensive extracellular remodeling and fibrosis in obesity (Weisberg, McCann et al. 2003, Nishimura, Manabe et al. 2009). Surprisingly, our transcriptomic analysis identified that pathways related to angiogenesis and fibrosis were enriched in the shiftwork group, albeit without pronounced obesity. As shown in Fig. 6A, when we compared gene expression in eWAT of normal controls at in ZT3 vs. ZT12, we found strong diurnal expression of ECM-angiogenesis related genes. In addition, parallel to the persistent induction of inflammatory response, the chronic shiftwork group also displayed robust up-regulation of ECM-angiogenic genes at ZT12 as compared to that of ZT3 with loss of the diurnal pattern of expression (Fig. 6A). A trend of augmented expression of this pathway was evident in the iWAT as well (Fig. 6B). We thus examined the extent of fibrosis in adipose depots using Masson’s trichrome staining that specifically visualizes collagen (Fig. 6C & 6D). Consistent with the findings of angiogenic and fibrotic response induced by shiftwork, typical blue collagen stain was scarce in the normal controls, whereas markedly pronounced staining with characteristic distribution within interstitia of surrounding blood vessels were frequently found in the chronic shiftwork group eWAT (Fig. 6C). Certain amount of weak collagen deposition was also seen within adipose tissue in regions that that were not associated with blood vessels. Similar to eWAT, accumulation of blood vessels together with more abundant collagen were detected in iWAT under the chronic shift condition (Fig. 6D), while some collagen deposition was not confined to areas surrounding the vessels. Thus, transcriptomic and histological findings indicated augmented ECM remodeling and angiogenic processes by shiftwork regimen, even without development of obesity in this model.
Figure 6.
Chronic shiftwork induces adipose tissue fibrotic response and angiogenic pathway. (A, B) Heatmap presentation of differentially-expressed genes involved in angiogenesis pathway in 6-month shiftwork and normal control groups at 8AM and 5PM in eWAT (A), and (B) 8AM samples in iWAT (n=3/group). Transcripts are annotated and expression levels shown according to color scale. (C, D) Representative images of Masson’s Trichrome staining of eWAT (C) and iWAT (D) in normal control and shiftwork groups. Scale bar: 100 μM.
3.7. Chronic shiftwork results in insulin resistance in adipose depots
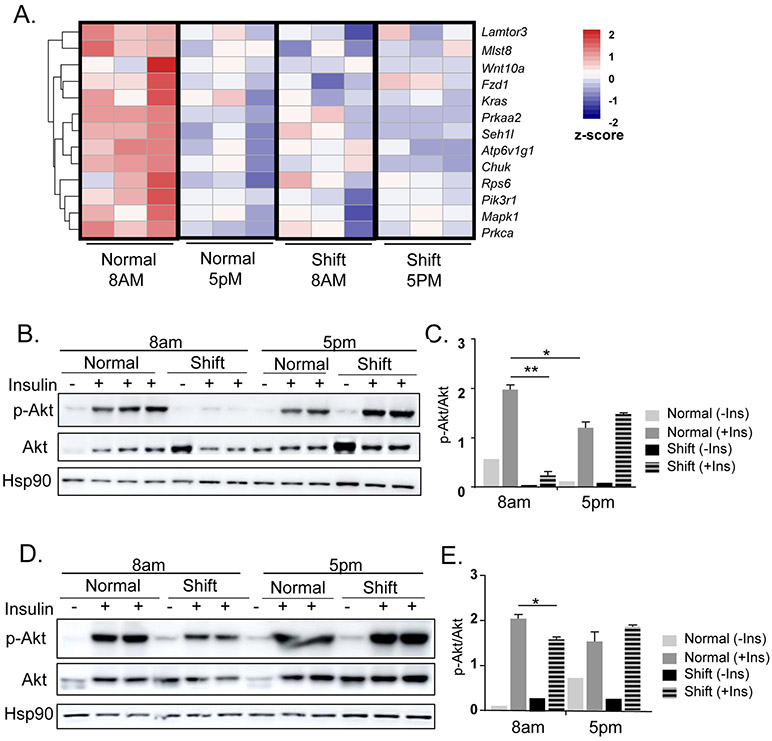
Adipose tissue inflammation and fibrosis is intimately linked with the development of insulin resistance (Crewe, An et al. 2017, Marcelin, Silveira et al. 2019), and loss of insulin sensitivity in fat depots significantly contributes to impaired whole-body glucose homeostasis. In visceral fat depot, RNA-Seq identified down-regulation of PI3K-mTOR pathway as a significantly affected process by the shiftwork regimen (Fig. 7A). Interestingly, contrary to the diurnal patterns of inflammatory, adipogenic or ECM-angiogenic pathways, a robust inhibition of mTOR signaling components was evident at ZT12 as compared to ZT3 (Fig. 7A). The shift group displayed consistently lower expression of mTOR pathway genes regardless of circadian time points, suggesting a potential dampening of mTOR activity induced by shiftwork. Based on the down-regulation of PI3K-mTOR pathway, we examined whether insulin sensitivity could be impaired in the adipose tissues of mice subjected to chronic shiftwork. In both eWAT and iWAT, shiftwork significantly impaired insulin signaling as assessed by Akt activation in response to acute insulin stimulation. Interestingly, as shown in Fig. 7B & 7C in eWAT, consistent with the robust mTOR pathways expression, Akt activation at ZT3 was significantly elevated than that of ZT12, indicating a daily variation of insulin sensitivity that was high in the early morning time. In addition, insulin stimulated robust AKT phosphorylation at ZT3 in normal controls, whereas the phosphorylation induced by insulin was nearly abolished in the shifted group. However, at ZT12, the Akt response to insulin was almost reversed in the shift groups with a tendency for increased phosphorylation that coincides with the dampened expression of the mTOR pathway in this cohort. Thus, the altered insulin sensitivity largely mirrors the gene expression regulation of mTOR components revealed by the transcriptomic analysis. In the iWAT, shiftwork also induced a diminished Akt response upon insulin stimulation at ZT3, although not as strongly affected as eWAT. Akt activation in response to insulin was comparable between two cohorts at ZT12 (Fig. 7D &7E). Together, these analyses revealed a diurnal regulation of insulin sensitivity in both adipose depots and chronic shift significantly impaired insulin signaling.
Figure 7.
Chronic shiftwork leads to insulin resistance in adipose depots. (A) Heatmap presentation of differentially-expressed genes in mTOR pathway in 6-month shiftwork and normal control groups at 8AM and 5PM in eWAT. (B-E) Insulin sensitivity as assessed by immunoblot analysis of insulin-stimulated Akt phosphorylation level normalized to total Akt at 8AM and 5PM in eWAT (B) with quantification (C), and in iWAT (D) with quantification (E). Values are expressed as p-Akt level normalized to total Akt as quantified by ImageJ. Hsp90 level shown as additional loading control. n=6/group, with 2-3 pooled samples for each lane. *: P≤0.05 or **: P≤0.01 by Student’s t test.
4. DISCUSSION
As a major site for energy storage, various aspects of adipose tissue function are controlled by circadian clock to coordinate with whole-body metabolic homeostasis (Shostak, Meyer-Kovac et al. 2013, Lekkas and Paschos 2019). Disruption of circadian clock regulation predispose to obesity and diabetes (Pan, Schernhammer et al. 2011, Roenneberg, Allebrandt et al. 2012, Laermans and Depoortere 2015). Despite the strong associated risk ratios detected among shiftworkers with metabolic disease, particularly type II diabetes, few studies to date examined this link in experimental settings. Using a chronic lighting-induced clock disruption regimen to mimic long-term shiftwork schedule, our study demonstrates a pronounced effect of chronic shiftwork on adipose tissue expansion and inflammation that leads to significant insulin resistance. Global transcriptomic profiling uncovered shiftwork-altered pathways that underlie adipocyte hypertrophy and impaired insulin signaling. Given the established link between clock disruption and the development of obesity and insulin resistance, perturbation of adipose tissue remodeling and insulin sensitivity could be a significant etiology underlying the current epidemic of metabolic disorders.
The most striking effect of chronic shift in adipose depots is adipocyte hypertrophy with accompanying inflammation and fibrosis. Development of obesity, often as a result of overnutrition, is characterized by extensive adipose tissue remodeling that involves adipocyte hypertrophy, inflammatory infiltration, lipid mobilization, and ECM remodeling (Crewe, An et al. 2017, Marcelin, Silveira et al. 2019). Interestingly, chronic shift induced these typical characteristics of overnutrition-induced obesity without any diet manipulation. RNA-seq analyses of adipose depots subjected to chronic shift revealed perturbed pathways implicated in adipose tissue remodeling, with enriched molecular processes encompassing the full spectrum of the pathological features of obesity. The effect of circadian disruption on obesity may depend on the specific type of circadian shift examined as well as the duration. The precise type and amount of clock disruption varies between studies with experimental conditions, such as constant lighting, dim light at night, phase advance or delay light shift, fixed shortening, lengthening or alternating day-night periods (Scheer, Hilton et al. 2009, Karatsoreos, Bhagat et al. 2011). Many large-scale epidemiological studies of shiftworkers that reveal increased risk for obesity and insulin resistance were not confined to a specific type of shiftwork in distinct populations, although the duration of the shiftwork correlated with the severity of adverse effects (van Amelsvoort, Schouten et al. 1999, Parkes 2002, Pan, Schernhammer et al. 2011, Buxton, Cain et al. 2012). Shifts in environmental lighting affect both the central clock and peripheral adipose tissue clocks (Lekkas and Paschos 2019), while central clock disruption alters feeding behavior and locomotor activity to impair energy balance (Karatsoreos et al., 2011,McHill et al., 2014,Kolbe, Brehm and Oster, 2019). Dysregulation of clock metabolic output pathways may collectively contribute to obesity, with misalignment of the central and peripheral clock circuits altering metabolic homeostasis and energy balance (Karatsoreos, Bhagat et al. 2011, McGowan and Coogan 2013). Given that the specific schedules within the shiftwork populations can be quite variable, how to best model and quantitatively measure its metabolic consequence remains to be critical to better understand and minimize its metabolic disease consequences (Rea and Figueiro 2014, Tsang, Astiz et al. 2017).
Shift-induced adipose hypertrophy resulted in prominent accumulation of macrophage infiltration, a hallmark of adipose inflammation (Weisberg, McCann et al. 2003, Nishimura, Manabe et al. 2009). Notably, chronic shiftwork can induce a systemic inflammatory state with documented heightened susceptibility to infections and inflammatory diseases (Castanon-Cervantes, Wu et al. 2010, Adams, Castanon-Cervantes et al. 2013, Fonken, Weil et al. 2013). Key components of the molecular clock network directly regulate inflammation and immune functions (Carter, Durrington et al. 2016, Scheiermann, Gibbs et al. 2018). Core clock genes, including both positive and negative regulators, including Bmal1, CLOCK, Rev-erbα and Per/Cry, are known to modulate innate and adaptive immune responses (Scheiermann, Gibbs et al. 2018). Macrophage cytokine production is under the transcription control of Bmal1/CLOCK and Rev-erbα (Gibbs, Blaikley et al. 2012, Spengler, Kuropatwinski et al. 2012). Both B and T lymphocyte functions are modulated by Bmal1(Sun, Yang et al. 2006), while RORγ and Rev-erbα, (Yang, Pappu et al. 2008), the positive and negative transcription regulators of a RORE DNA-binding element respectively, are key players of Th17 lymphocyte development. Our study revealed robust time-of-the-day-dependent expression of immune response genes in adipose tissue. The induction of immune pathways at late afternoon as compared to early morning suggests a heightened immune function at the beginning of the activity cycle, potentially an anticipatory mechanism intrinsic to immune regulation. The dysregulation of macrophage and immune functions in adipose depots induced by chronic shift could be attributed to distinct mechanisms. Disrupted cell-autonomous clock control in immune cells may lead to inflammation, or the inflammatory response could be secondary to adipose expansion. Adipose tissue expansion requires extensive ECM remodeling with neovascularization leading to inflammation, while chronic inflammatory milieu in obesity further exacerbates ECM deposition and aberrant angiogenesis (Nishimura, Manabe et al. 2007, Chun 2012). Macrophage infiltration, tissue fibrosis and neovascularization are prominent features we identified in the shiftwork adipose depots. As indicated by the abundant crown-like structures composed of macrophages surrounding necrotic adipocytes, shiftwork adipose depots display pronounced macrophage infiltration. Macrophage-induced inflammatory signals are known to stimulate adipocyte lipid release, activate interstitial cell types and induce extracellular matrix remodeling and deposition in adipose depots (Marcelin, Silveira et al. 2019). These processes, all consequences of circadian clock dysregulation, could intertwine in the context of chronic shift-induced obesity to contribute to attenuate systemic metabolic homeostasis over time.
Our finding of the circadian time-dependent regulation of adipocyte cell differentiation processes and their persistent elevation in shiftwork groups provides a potential mechanism underlying adipocyte hypertrophy. Additionally, this is accompanied by up-regulation of lipid metabolic processes indicative of lipid flux to promote new adipocyte formation or hypertrophic adipocyte growth. We observed diurnal protein expression patterns of adipogenic factors, C/EBPβ and PPARγ, both being induced by shiftwork at ZT2. This diurnal regulation and induction by shift of adipogenic pathways support the notion that these are metabolic outputs controlled by the clock circuit. Notably, key transcription regulators of the core clock circuit, including Bmal1, Rev-erbα and Per2, were known to regulate adipogenic differentiation (Grimaldi, Bellet et al. 2010, Guo, Chatterjee et al. 2012, Nam, Chatterjee et al. 2015). In addition, lipid mobilization and storage in adipose depots demonstrate clear circadian control (Shostak, Meyer-Kovac et al. 2013). The pronounced adipose expansion induced by chronic shift we observed could be a result of clock gene dysregulation on adipogenic and lipid metabolic processes. However, as circadian shift alters both the central and peripheral clock network, metabolic changes in adipose tissue likely reflects the consequence of dysfunctional adipose tissue clock and SCN input to adipose depots. Precise dissection of metabolic and energy homeostasis under shiftwork conditions in future studies may provide insights into the distinct mechanisms that collectively drive adipose expansion.
Consistent with the well-recognized strong association between clock disruption and the development of Type II diabetes (Pan, Schernhammer et al. 2011), we found significantly impaired adipose tissue insulin signaling in mice undergone 6 months of shiftwork regimen. This adipose depot insulin resistance induced by clock disruption in our chronic shiftwork model may shed light on the mechanisms contributing to Type II diabetes in a modern workforce with circadian misalignment. Two mechanisms we examined in this study could underly the insulin insensitivity in the shiftwork cohort. Circadian variations of the mTOR signaling pathway was evident in eWAT, and chronic shift strongly suppressed this pathway. In addition, impaired insulin signaling could be a consequence of the obesity and associated inflammation in the adipose depots. It is also worth noting, that the phospho-Akt in response to insulin was significantly higher in both eWAT and iWAT at early morning hours that corresponds to increased mTOR signaling pathway gene expression. This finding may have specific clinical implications at the dosing of insulin in diabetic patients may vary due to this intrinsic time-of-the-day variation. Our findings are in line with previous reports of the circadian control involvement in the mTOR signaling pathways, although most previous studies are largely confined to liver (Cao, Robinson et al. 2013, Lipton, Yuan et al. 2015, Ramanathan, Kathale et al. 2018). The adipose tissue insulin resistance as a result of chronic shift schedule could be a significant contributor to the well-documented link between systemic insulin resistance and clock misalignment in large-scale epidemiological studies.
In our modern lifestyle, clock disruption occurs at an alarming frequency when artificial lighting or sleep/activity schedule is misaligned with the endogenous clock cycles. This may constitute a potentially wide-spread etiology underlying the current epidemics of metabolic diseases (van Amelsvoort, Schouten et al. 1999, Pan, Schernhammer et al. 2011, Vetter, Devore et al. 2015). Our current study using an experimental shiftwork paradigm uncovers the adipose tissue as a key site that contributes to the development of obesity and insulin resistance. Better understandings of the precise circadian etiologies underlying the current epidemics of obesity may lead to new targeted interventions to modulate clock function for prevention or treatment.
Supplementary Material
Highlights.
Chronic shiftwork in mice induced marked adipocyte hypertrophy of visceral and subcutaneous depots.
Chronic shift led to macrophage infiltration with up-regulated inflammatory pathways.
Transcriptomic profiling revealed enriched processes characteristic of obesity in chronic shiftwork.
Diurnal regulation of adipogenic and lipid metabolic pathways were altered in shift-induced adipose expansion.
Chronic shiftwork impaired insulin signaling and suppressed mTOR pathway in adipose depots.
ACKNOWLEDGEMENTS
We thank the Shared Resources Core Facility of City of Hope for their expert technical assistance on RNA-Seq analyses. K. M is a faculty member supported by the NCI-designated Comprehensive Cancer Center at the City of Hope National Cancer Center. This project was supported by grants from National Institute of Health 1R01DK112794, and American Heart Association 17GRNT33370012 to KM; and National Institute of Health grant DK097160-01 to VY, American Heart Association grant 18TPA34230103 to AP, and National Institute of occupational Health and Safety R01OH010668 to MGF.
Non-standard Abbreviations:
- Bmal1
Brain and Muscle Arnt-like Protein 1
- CLOCK
Circadian Locomotor Output Cycle
- ECM
extracellular matrix
- eWAT
epidydimal white adipose tissue
- iWAT
inguinal white adipose tissue
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
I certify that neither I nor my co-authors have a conflict of interest as described above that is relevant to the subject matter or materials included in this work.
Reference
- Adams KL, Castanon-Cervantes O, Evans JA and Davidson AJ (2013). "Environmental circadian disruption elevates the IL-6 response to lipopolysaccharide in blood." J Biol Rhythms 28(4): 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT and Huber W (2015). "HTSeq--a Python framework to work with high-throughput sequencing data." Bioinformatics 31(2): 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM and Sherlock G (2000). "Gene ontology: tool for the unification of biology. The Gene Ontology Consortium." Nat Genet 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J and Takahashi JS (2010). "Circadian integration of metabolism and energetics." Science 330(6009): 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton OM, Cain SW, O'Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA and Shea SA (2012). "Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption." Sci Transl Med 4(129): 129ra143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S and Sonenberg N (2013). "Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling." Neuron 79(4): 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SJ, Durrington HJ, Gibbs JE, Blaikley J, Loudon AS, Ray DW and Sabroe I (2016). "A matter of time: study of circadian clocks and their role in inflammation." J Leukoc Biol 99(4): 549–560. [DOI] [PubMed] [Google Scholar]
- Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, Besing RC, Menaker M, Gewirtz AT and Davidson AJ (2010). "Dysregulation of inflammatory responses by chronic circadian disruption." J Immunol 185(10): 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TH (2012). "Peri-adipocyte ECM remodeling in obesity and adipose tissue fibrosis." Adipocyte 1(2): 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewe C, An YA and Scherer PE (2017). "The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis." J Clin Invest 127(1): 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC and Lempicki RA (2003). "DAVID: Database for Annotation, Visualization, and Integrated Discovery." Genome Biol 4(5): P3. [PubMed] [Google Scholar]
- Dibner C, Schibler U and Albrecht U (2010). "The mammalian circadian timing system: organization and coordination of central and peripheral clocks." Annu Rev Physiol 72: 517–549. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013). "STAR: ultrafast universal RNA-seq aligner." Bioinformatics 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken LK, Weil ZM and Nelson RJ (2013). "Mice exposed to dim light at night exaggerate inflammatory responses to lipopolysaccharide." Brain Behav Immun 34: 159–163. [DOI] [PubMed] [Google Scholar]
- Fontaine C, Dubois G, Duguay Y, Helledie T, Vu-Dac N, Gervois P, Soncin F, Mandrup S, Fruchart JC, Fruchart-Najib J and Staels B (2003). "The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation." J Biol Chem 278(39): 37672–37680. [DOI] [PubMed] [Google Scholar]
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW and Loudon AS (2012). "The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines." Proc Natl Acad Sci U S A 109(2): 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T and Sassone-Corsi P (2010). "PER2 controls lipid metabolism by direct regulation of PPARgamma." Cell Metab 12(5): 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, Minze LJ, Hsueh W and Ma K (2012). "The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway." FASEB J 26(8): 3453–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M and Goto S (2000). "KEGG: kyoto encyclopedia of genes and genomes." Nucleic Acids Res 28(1): 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH and McEwen BS (2011). "Disruption of circadian clocks has ramifications for metabolism, brain, and behavior." Proc Natl Acad Sci U S A 108(4): 1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laermans J and Depoortere I (2015). "Chronobesity: role of the circadian system in the obesity epidemic." Obes Rev. [DOI] [PubMed] [Google Scholar]
- Lee J, Moulik M, Fang Z, Saha P, Zou F, Xu Y, Nelson DL, Ma K, Moore DD and Yechoor VK (2013). "Bmal1 and beta-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced beta-cell failure in mice." Mol Cell Biol 33(11): 2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkas D and Paschos GK (2019). "The circadian clock control of adipose tissue physiology and metabolism." Auton Neurosci 219: 66–70. [DOI] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM and Sahin M (2015). "The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation." Cell 161(5): 1138–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Xiong X, Nam D, Yechoor V and Ma K (2020). "SRF-MRTF signaling suppresses brown adipocyte development by modulating TGF-beta/BMP pathway." Mol Cell Endocrinol 515: 110920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W and Anders S (2014). "Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2." Genome Biol 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin G, Silveira ALM, Martins LB, Ferreira AV and Clement K (2019). "Deciphering the cellular interplays underlying obesity-induced adipose tissue fibrosis." J Clin Invest 129(10): 4032–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan NM and Coogan AN (2013). "Circadian and behavioural responses to shift worklike schedules of light/dark in the mouse." J Mol Psychiatry 1(1): 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHill AW, Melanson EL, Higgins J, Connick E, Moehlman TM, Stothard ER and Wright KP Jr. (2014). "Impact of circadian misalignment on energy metabolism during simulated nightshift work." Proc Natl Acad Sci U S A 111(48): 17302–17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D, Chatterjee S, Yin H, Liu R, Lee J, Yechoor VK and Ma K (2015). "Novel Function of Rev-erbalpha in Promoting Brown Adipogenesis." Sci Rep 5: 11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D, Guo B, Chatterjee S, Chen MH, Nelson D, Yechoor VK and Ma K (2015). "The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways." J Cell Sci 128(9): 1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, Yoshimura K, Kadowaki T and Nagai R (2009). "CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity." Nat Med 15(8): 914–920. [DOI] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Nagasaki M, Hosoya Y, Yamashita H, Fujita H, Ohsugi M, Tobe K, Kadowaki T, Nagai R and Sugiura S (2007). "Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels." Diabetes 56(6): 1517–1526. [DOI] [PubMed] [Google Scholar]
- Otway DT, Frost G and Johnston JD (2009). "Circadian rhythmicity in murine preadipocyte and adipocyte cells." Chronobiol Int 26(7): 1340–1354. [DOI] [PubMed] [Google Scholar]
- Pan A, Schernhammer ES, Sun Q and Hu FB (2011). "Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women." PLoS Med 8(12): e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS and Hogenesch JB (2002). "Coordinated transcription of key pathways in the mouse by the circadian clock." Cell 109(3): 307–320. [DOI] [PubMed] [Google Scholar]
- Parkes KR (2002). "Shift work and age as interactive predictors of body mass index among offshore workers." Scand J Work Environ Health 28(1): 64–71. [DOI] [PubMed] [Google Scholar]
- Ramanathan C, Kathale ND, Liu D, Lee C, Freeman DA, Hogenesch JB, Cao R and Liu AC (2018). "mTOR signaling regulates central and peripheral circadian clock function." PLoS Genet 14(5): e1007369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea MS and Figueiro MG (2014). "Quantifying light-dependent circadian disruption in humans and animal models." Chronobiol Int 31(10): 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T, Allebrandt KV, Merrow M and Vetter C (2012). "Social jetlag and obesity." Curr Biol 22(10): 939–943. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS and Shea SA (2009). "Adverse metabolic and cardiovascular consequences of circadian misalignment." Proc Natl Acad Sci U S A 106(11): 4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann C, Gibbs J, Ince L and Loudon A (2018). "Clocking in to immunity." Nat Rev Immunol 18(7): 423–437. [DOI] [PubMed] [Google Scholar]
- Schibler U and Sassone-Corsi P (2002). "A web of circadian pacemakers." Cell 111(7): 919–922. [DOI] [PubMed] [Google Scholar]
- Shostak A, Meyer-Kovac J and Oster H (2013). "Circadian regulation of lipid mobilization in white adipose tissues." Diabetes 62(7): 2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV and Antoch MP (2012). "Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription." Proc Natl Acad Sci U S A 109(37): E2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang Z, Niu Z, Peng J, Li Q, Xiong W, Langnas AN, Ma MY and Zhao Y (2006). "MOP3, a component of the molecular clock, regulates the development of B cells." Immunology 119(4): 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017). "Transcriptional architecture of the mammalian circadian clock." Nat Rev Genet 18(3): 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AH, Astiz M, Leinweber B and Oster H (2017). "Rodent Models for the Analysis of Tissue Clock Function in Metabolic Rhythms Research." Front Endocrinol (Lausanne) 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS and Bass J (2005). "Obesity and metabolic syndrome in circadian Clock mutant mice." Science 308(5724): 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amelsvoort LG, Schouten EG and Kok FJ (1999). "Duration of shiftwork related to body mass index and waist to hip ratio." Int J Obes Relat Metab Disord 23(9): 973–978. [DOI] [PubMed] [Google Scholar]
- Vetter C, Devore EE, Ramin CA, Speizer FE, Willett WC and Schernhammer ES (2015). "Mismatch of Sleep and Work Timing and Risk of Type 2 Diabetes." Diabetes Care 38(9): 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL and Ferrante AW Jr. (2003). "Obesity is associated with macrophage accumulation in adipose tissue." J Clin Invest 112(12): 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zvonic S, Floyd ZE, Kilroy G, Goh BC, Hernandez TL, Eckel RH, Mynatt RL and Gimble JM (2007). "Induction of circadian gene expression in human subcutaneous adipose-derived stem cells." Obesity (Silver Spring) 15(11): 2560–2570. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM and Dong C (2008). "T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma." Immunity 28(1): 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL and Gimble JM (2006). "Characterization of peripheral circadian clocks in adipose tissues." Diabetes 55(4): 962–970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.