Abstract
Among 3302 persons tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by BinaxNOWTM and reverse transcription polymerase chain reaction (RT-PCR) in a community setting, rapid assay sensitivity was 100%/98.5%/89% using RT-PCR cycle thresholds of 30, 35, and no threshold. The specificity was 99.9%. Performance was high across ages and those with and without symptoms. Rapid resulting permitted immediate public health action.
Keywords: community-based SARS-CoV-2 testing, asymptomatic SARS-CoV-2 infection
Breaking severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) community transmission chains requires rapid identification and isolation of infectious persons. Up to 40% of infected persons may not have symptoms, despite harboring high levels of virus [1]. Further, standard testing models pose multiple barriers to the effective use of testing for epidemic control, including testing access restricted to symptomatic persons, difficult appointment scheduling, long turnaround times, and structural barriers including health insurance, monolingual services, and location of testing sites far from communities most affected. Deploying rapid antigen tests with high field performance through the use of community-based test-and-respond models [2] could address these barriers and increase the identification of the most infectious persons. Importantly, compared with a standard reverse transcription polymerase chain reaction (RT-PCR) assay, use of these tests could rapidly permit identification and isolation of persons with high levels of virus, disrupting forward transmission chains [3].
We evaluated the Abbott BinaxNOWTM coronavirus disease 2019 (COVID-19) antigen card rapid assay performance for detection of persons with high levels of virus and measured the time to isolation in a community walkup test-and-respond program.
METHODS
Study Setting and Procedures
We conducted this study through an academic, community (Latino Task Force) and public health department partnership (Unidos en Salud). We offered testing at a plaza under tents in an urban commercial transport hub in the Mission neighborhood in San Francisco, a setting of ongoing community transmission, predominantly among Latinx persons. Community workers conducted door-to-door mobilization in 3 census tracts surrounding the testing site 4 days before testing. Persons of all ages, with or without symptoms, were registered onsite. After consent, trained community volunteers conducted a brief survey that included demographic information and COVID-19 symptoms. Certified laboratory assistants collected bilateral anterior nasal swab for BinaxNOW (cards provided by State of California Department of Public Health) according to manufacturer instructions, immediately followed by a separate bilateral swab for RT-PCR. BinaxNOW results were read on site by certified technician readers [4, 5]. We returned positive rapid antigen test results via secure messaging within an hour of testing and a follow-up phone call occurred within 2 hours. Staff provided counseling and offered a city-sponsored hotel stay for isolation. Persons choosing to isolate at home had immediate same-day access to home services, including health education and food delivery, administered through a community-led outreach program [6]. Health department contact tracing was initiated immediately on return of a positive BinaxNOW result.
RT-PCR was completed by RenegadeBio using RenegadeXPTM technology. Anterior nares swabs were collected into proprietary viral transport media, then lysed. Lysate was transferred directly to a multiplex RT-PCR reaction with primers/probes for the nucleoprotein gene of SARS-CoV-2. Positive results were confirmed by the standard US Centers for Disease Control and Prevention methodology using Qiagen viral RNA purification kits and singleplex RT-PCR detection of the nucleoprotein gene.
BinaxNOW Assay sensitivity and specificity with 95% confidence intervals were calculated using RT-PCR cycle thresholds (Ct) below 30 and 35 (corresponding to high viral levels associated in vitro with virus viability) [7–9]. Time to reporting was calculated from time of registration to time of test results notification. Time to isolation was calculated from symptom onset for those persons who were symptomatic before or at the time of testing.
Ethics Statement
The University of California San Francisco Committee on Human Research determined that the study met criteria for public health surveillance. All participants provided informed consent for dual testing.
RESULTS
We tested 3302 persons over 6 days between November 22 and December 1, 2020; 99 were aged <13 years, 110 aged 13 to 18 years, and 3093 aged >18 years. Participants were 45.4% female and 53.0% male. Reported ethnicity was 65.6% Latinx, 9.2% Asian, 16.9% White, 1.6% American Indian, and 2.5% Black. Of all persons tested, 30.9% self-reported possible COVID-19 symptoms. At this site, equipped with 3 testing tents each with 4 technicians and 1 data entry volunteer, we were able to test approximately 100 persons per hour.
There were 237 persons overall who were RT-PCR positive (7.2% prevalence), and 211 (6.4%) persons who were also rapid test positive. Postitive RT-PCR positive prevalence was 19/99 (19.4%) among children <13 years of age, and 16/110 (14.5%) among teens 13 to 18 years of age. Ninety-five RT-PCR(+) persons (40.1%) were asymptomatic and 7 (3.0%) had a symptom that started >7 days before testing.
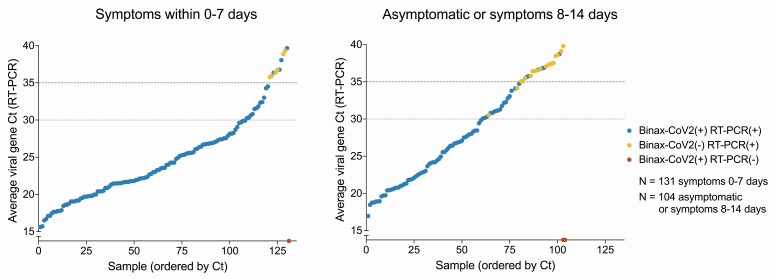
The BinaxNOW test exhibited high sensitivity and specificity for persons with high levels of virus (Ct <30 or <35), both overall and stratified by age and presence of symptoms (Table 1). Sensitivity using a Ct cutoff of 30 was 100% (95% confidence interval [CI]: 97.9–100) for the full study population, 100% (95% CI: 73.5–100) among persons <13 years of age, and 100% (95% CI: 73.5-100) among persons 13 to 18 years of age. Among 102 persons who were asymptomatic or whose symptom onset was >7 days before testing, sensitivity for a Ct cutoff of 30 was 100% (95% CI: 94-100). Persons with and without symptoms exhibited a similar range of Ct levels by RT-PCR (Figure 1). Three individuals were rapid test positive and RT-PCR test negative, resulting in a BinaxNOWTM false positive rate of 0.1% (3/3065); 1 had symptoms (cough). Overall test specificity was 99.9% (95% CI: 99.7-100).
Table 1.
Sensitivity and Specificity of BinaxNOW Stratified by Age and Symptoms
| Populations | BinaxNOW Performance | All | Symptom Onset Within 7 Daysa | Asymptomatic or Symptom Onset >7 Days Ago |
|---|---|---|---|---|
| All ages (N = 3302) Value (95% CI) |
Ct = 30 cutoff | |||
| Sensitivity | 100% (171/171, 95% CI: 97.9–100) | 100% (108/108, 95% CI: 96.6–100) | 100% (60/60, 95% CI: 94–100) | |
| Specificity | 98.6% (3088/3131, 95% CI: 98.2–99) | 97% (546/563, 95% CI: 95.2–98.2) | 98.9% (2317/2342, 95% CI: 98.4–99.3) | |
| Ct = 35 cutoff | ||||
| Sensitivity | 98.5% (201/204, 95% CI: 95.8–99.7) | 100% (120/120, 95% CI: 97–100) | 97.5% (77/79, 95% CI: 91.2–99.7) | |
| Specificity | 99.6% (3085/3098, 95% CI: 99.3–99.8) | 99.1% (546/551, 95% CI: 97.9–99.7) | 99.7% (2315/2323, 95% CI: 99.3–99.9) | |
| No CT cutoff | ||||
| Sensitivity | 89% (211/237, 95% CI: 84.3–92.7) | 95.4% (124/130, 95% CI: 90.2–98.3) | 81.4% (83/102, 95% CI: 72.4–88.4) | |
| Specificity | 99.9% (3062/3065, 95% CI: 99.7–100) | 99.8% (540/541, 95% CI: 99–100) | 99.9% (2298/2300, 95% CI: 99.7–100) | |
| Ages < 13 years (N = 99) Value (95% CI) |
Ct = 30 cutoff | |||
| Sensitivity | 100% (12/12, 95% CI: 73.5–100) | 100% (3/3, 95% CI: 29.2–100) | 100% (9/9, 95% CI: 66.4–100) | |
| Specificity | 96.6% (84/87, 95% CI: 90.3–99.3) | 91.7% (11/12, 95% CI: 61.5–99.8) | 97.1% (68/70, 95% CI: 90.1–99.7) | |
| Ct = 35 cutoff | ||||
| Sensitivity | 93.3% (14/15, 95% CI: 68.1–99.8) | 100% (3/3, 95% CI: 29.2–100) | 91.7% (11/12, 95% CI: 61.5–99.8) | |
| Specificity | 98.8% (83/84, 95% CI: 93.5–100) | 91.7% (11/12, 95% CI: 61.5–99.8) | 100% (67/67, 95% CI: 94.6–100) | |
| No Ct cutoff | ||||
| Sensitivity | 78.9% (15/19, 95% CI: 54.4–93.9) | 80% (4/5, 95% CI: 28.4–99.5) | 78.6% (11/14, 95% CI: 49.2–95.3) | |
| Specificity | 100% (80/80, 95% CI: 95.5–100) | 100% (10/10, 95% CI: 69.2–100) | 100% (65/65, 95% CI: 94.5–100) | |
| Ages 13-18 years (N = 110) Value (95% CI) |
Ct = 30 cutoff | |||
| Sensitivity | 100% (12/12, 95% CI: 73.5–100) | 100% (8/8, 95% CI: 63.1–100) | 100% (4/4, 95% CI: 39.8–100) | |
| Specificity | 96.9% (95/98, 95% CI: 91.3–99.4) | 100% (13/13, 95% CI: 75.3–100) | 96.1% (73/76, 95% CI: 88.9–99.2) | |
| Ct = 35 cutoff | ||||
| Sensitivity | 100% (14/14, 95% CI: 76.8–100) | 100% (8/8, 95% CI: 63.1–100) | 100% (6/6, 95% CI: 54.1–100) | |
| Specificity | 99% (95/96, 95% CI: 94.3–100) | 100% (13/13, 95% CI: 75.3–100) | 98.6% (73/74, 95% CI: 92.7–100) | |
| No Ct cutoff | ||||
| Sensitivity | 93.8% (15/16, 95% CI: 69.8–99.8) | 100% (8/8, 95% CI: 63.1–100) | 87.5% (7/8, 95% CI: 47.3–99.7) | |
| Specificity | 100% (94/94, 95% CI: 96.2–100) | 100% (13/13, 95% CI: 75.3–100) | 100% (72/72, 95% CI: 95–100) |
Abbreviations: CI, confidence interval; Ct, cycle threshold.
a Symptom onset date missing for 229 persons.
Figure 1.
RT-PCR Ct values and BinaxNOW rapid antigen test results of participants, stratified according to COVID-19 symptoms. Average viral Ct values of all individuals with positive RT-PCR and/or rapid antigen test results (N = 245 total) plotted in ascending order of Ct. Each point represents 1 individual. Blue points are individuals whose samples were positive for both rapid antigen test (BinaxNOW) and RT-PCR test. Yellow circles represent individuals who were RT-PCR positive, but rapid antigen test negative. Red circles represent individuals with a positive rapid antigen test and a negative RT-PCR test result. Abbreviations: COVID-19, coronavirus disease 2019; Ct, cycle threshold; RT-PCR, reverse transcription polymerase chain reaction.
For persons with a positive rapid antigen test, the median time from onsite registration to electronic results notification (N = 211) was 62 minutes (interquartile range: 47–82 minutes). Phone calls followed within 1 hour. Among symptomatic persons with a positive rapid antigen test (N = 134), the median time from symptom onset to isolation using BinaxNOW antigen test results was 3 days (interquartile range: 2–5 days).
DISCUSSION
SARS-CoV-2 pandemic control calls for fast, low-barrier, high-performing field assays accessible to people who will not otherwise be tested or who will receive results too late for results to make a difference. The US government has purchased 150 million BinaxNOW cards, yet their use to date has been limited because of gaps in information about performance and assessment of public health activation. Our data show that these tests are readily deployed in a field setting at scale for children and adults, can rapidly identify persons with high levels of virus including those who are asymptomatic, and can lead to immediate public health action.
A major benefit of using this high-performing rapid test was the speed with which results were returned (approximately 1 hour from walkup registration to return). This permitted immediate public health action for persons infected with high levels of virus, who are most likely to be infectious [3, 10, 11]. Upon receipt of a positive rapid test result, we activated an isolation protocol offering city sponsored hotels or home isolation with supportive services. In addition, contact tracing was initiated 24 to 48 hours earlier than would have been possible through routine city-sponsored RT-PCR testing. The use of this technology further allowed rapid mass screening in an outside setting easily accessed by communities at high risk of ongoing transmission. This test is much less costly than RT-PCR and does not require a machine to read.
Integration of rapid antigen testing within community-based test and respond initiatives could contribute to reduced transmission via several mechanisms, including more complete and earlier detection of infectious persons made possible by increased access to low-barrier testing. Even without these benefits, however, the reduction in turnaround time alone afforded by BinaxNOW (approximately 1 hour) compared with an RT-PCR turnaround of 4 days could potentially eliminate 4 highly infectious days that would otherwise be spent out of isolation (of a typical 10-day maximum potential isolation period for nonhospitalized patients) [3, 10].
We found high sensitivity and specificity for the BinaxNOW assay, including in asymptomatic persons and children. These results expand on, and are concordant with, our previous report which found a BinaxNOW detection level of ~2 × 104 viral RNA copies based on titration experiments [12]. Heterogeneity in the relationship between Ct and viral load across RT-PCR platforms complicates direct comparisons of raw Ct values; however, we find BinaxNOW reliably detects persons with low Ct, correlating to high viral load. Rapid tests may miss individuals at the earliest rise in virus levels, a limitation that can be addressed through repeat rapid testing [3, 10]. Rapid tests may also miss the latter end of the viral dynamic curve (which can last for weeks), a period during which virus levels are low and a person is not thought to be infectious; some have suggested the lower sensitivity of the assay during this period could reduce hardship resulting from unnecessary isolation.
SARS-CoV-2 RT-PCR testing remains the gold standard for diagnosis. Even with a rapid test specificity of 99.9%, false positives will account for 2% or less of total BinaxNOW positives when SARS-CoV-2 prevalence is above 5%. For populations with a 2% SARS-CoV-2 prevalence, 5.1% of total BinaxNOW positives would be false positives. We used these tests in a high-prevalence community setting, where RT-PCR confirmation may not be required outside the research context. In other settings with lower prevalence, confirmatory RT-PCR would be required, particularly among persons without symptoms or exposures.
Our low-barrier testing model incorporating the rapid BinaxNOW assay and linked with supportive follow-up services could identify more infectious persons faster, decrease the time to isolation, and interrupt transmission chains. As a national vaccine rollout is implemented, strategic testing strategies remain a key part of the public health response.
Notes
Acknowledgments. We thank Bevan Dufty and the BART team, Supervisor Hillary Ronen, Mayor London Breed, Dr. Grant Colfax, Dr. Naveena Bobba, Dr. Jonathan Fuchs, Dr. Darpun Sachdev and the San Francisco Department of Public Health, Salu Ribeiro and Bay Area Phlebotomy and Laboratory services, Craig Rouskey and RenegadeBio, PrimaryBio COVID testing platform, and our dedicated community ambassadors and volunteers. We would also like to thank the California Department of Public Health for their generous donation of BinaxNOW COVID-19 Ag cards for the study.
Financial support. Funding for this study was provided by the University of California San Francisco, Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation, a private donor, the Chan Zuckerberg Initiative, and the National Institutes of Health [UM1AI069496].
Potential conflicts of interest. Dr. Havlir reports nonfinancial support from Abbott, outside the submitted work; none of the other authors has any potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med 2020; 173:362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chamie G, Marquez C, Crawford E, et al. SARS-CoV-2 community transmission disproportionately affects Latinx population during shelter-in-place in San Francisco. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larremore DB, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv 2020. doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unidos en Salud. Binax training resources. Available at: https://unitedinhealth.org/binax-training. Accessed 17 December 2020.
- 5. BinaxNOWTM COVID-19 Ag card and NAVICATM App set-up and training. Available at: https://www.globalpointofcare.abbott/en/support/product-installation-training/navica-brand/navica-binaxnow-ag-training.html. Accessed 17 December 2020.
- 6. Kerkhoff AD, Sachdev D, Mizany S, et al. Evaluation of a novel community-based COVID-19 ‘test-to-care’ model for low-income populations. PLoS One 2020; 15:e0239400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 2020; 39:1059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullard J, Dust K, Funk D, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020; 25:2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity - a strategy for containment. N Engl J Med 2020; 383:e120. [DOI] [PubMed] [Google Scholar]
- 11. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2020. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pilarowski G, Lebel P, Sunshine S, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. medRxiv 2020. doi: 10.1101/2020.11.02.20223891 [DOI] [PMC free article] [PubMed] [Google Scholar]