Abstract
The Novel Coronavirus Disease 2019 (COVID-19) has become an international public health emergency, which poses the most serious threat to the human health around the world. Accumulating evidences have shown that the new coronavirus could not only infect human beings, but also can infect other species which might result in the cross-species infections. In this research, 1056 ACE2 protein sequences are collected from the NCBI database, and 173 species with >60% sequence identity compared with that of human beings are selected for further analysis. We find 14 polar residues forming the binding interface of ACE2/2019-nCoV-Spike complex play an important role in maintaining protein–protein stability. Among them, 8 polar residues at the same positions with that of human ACE2 are highly conserved, which ensure its basic binding affinity with the novel coronavirus. 5 of other 6 unconserved polar residues (positions at human ACE2: Q24, D30, K31, H34 and E35) are proved to have an effect on the binding patterns among species. We select 21 species keeping close contacts with human beings, construct their ACE2 three-dimensional structures by Homology Modeling method and calculate the binding free energies of their ACE2/2019-nCoV-Spike complexes. We find the ACE2 from all the 21 species possess the capabilities to bind with the novel coronavirus. Compared with the human beings, 8 species (cow, deer, cynomys, chimpanzee, monkey, sheep, dolphin and whale) present almost the same binding abilities, and 3 species (bat, pig and dog) show significant improvements in binding affinities. We hope this research could provide significant help for the future epidemic detection, drug and vaccine development and even the global eco-system protections.
Introduction
The Coronavirus disease 2019 (COVID-19) caused by the virus SARS-CoV-2 has become an unprecedented international pandemic in our human history. According to the World Health Organization (WHO), as of 30 July 2020, more than 17 million COVID-19 cases have been reported in more than 200 countries and resulted in 667 thousand deaths. The 2019-nCoV coronavirus, together with Middle East Respiratory Syndrome coronavirus (MERS-CoV) [1] and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) [2], etc., can cross the species barriers and emerge as highly contagious virus.
In current reported cases, animals (camel, cat and bats) can serve as hosts of coronavirus, causing transmission between animals and human, which poses a greater threat to the public health and even the global ecosystem. The MERS-CoV was identified as zoonotic virus that could be transmitted between species within mammals, and various studies have shown that humans can be infected through direct or indirect contacts with infected dromedary camels [3]. In February 2004, the Chinese chrysanthemum bat was recognized as an intermediate host of SARS-like coronaviruses [4].
Now, various researches for finding the original coronavirus 2019-nCoV infecting sources have been conducted [5,6], and some species including Paguma larvata, Nyctereutes procyonoides [7], Bat [8,9,10] have been identified as suspect of the primary hosts of the novel coronavirus. The preliminary assessment indicated that manis might be the intermediate host of virus spreading to human beings by analyzing the evolutionary tree of 2019-nCoV coronavirus [11]. In addition, pets can contract certain types of coronaviruses, such as the canine respiratory coronavirus and the novel coronavirus, COVID-19. Two pet dogs, one in Hong Kong and one in Belgium, have been tested positive for COVID-19, and both of these dogs lived indoors with COVID-19 positive owners. Local health officials characterized the two cases of dogs as likely to be cases of human-to-animal transmission [12,13]. Furthermore, several tigers and lions at the zoo showed symptoms including dry cough, wheezing, and lack of appetite. All of the big cats with these symptoms at the zoo are believed to have been infected by a zoo employee who showed signs of COVID-19 [14]. The virus can transmit in cats via respiratory droplets [15]. SARS-CoV-2 even caused more severe interstitial pneumonia in old monkeys than that in young monkeys [16]. The latest studies suggested that the Ganges River, Macaca Mulatta monkeys, chicken, duck and mices can be infected with COVID-19 in different severity [17,18, 19, 20, 21, 22].
As of today, the precise hosts and primary infecting sources of 2019-nCoV still remained unclear. However, the cross-species infections between human beings and animals may have already occurred, as the first reported patient was found to have close contact with livestock. Moreover, novel coronavirus can mainly be transmitted via small respiratory droplets, aerosols, contacts and air, thus the other species are likely infected by COVID-19.
These emergency situations mentioned above urge us to discover species owning highly cross-species infection properties quickly, systematically and accurately and take more powerful measurements to prevent the spread of the COVID-19. To test which are the potentially infected animals and find what plays key roles in binding with coronavirus 2019-nCoV-Spike, we collect 1056 ACE2 protein sequences from the NCBI database and get 173 species which have more than 60% sequence identity of ACE2 proteins compared with that of human beings. We find that 14 polar residues at the same position on that of human ACE2 play major roles in forming the binding interface with coronavirus 2019-nCoV-Spike. Among them, 8 polar residues are highly conserved, which ensure its basic binding affinity with the novel coronavirus. 5 of the other 6 are unconserved polar residues which have different degrees of influences on the binding patterns among species. Finally, we select 21 species keeping close contacts with human beings for detailed analysis. For these 21 species, we construct their ACE2 three-dimensional structures, comparing the binding interfaces between their ACE2 proteins and Spike region of the novel coronavirus. Then we calculate the binding free energies of their ACE2/2019-nCoV-Spike complexes by using molecular dynamics simulation. All of the 21 species possess binding capabilities to the novel coronavirus.
Results
Key polar residues of ACE2 binding with coronavirus 2019-nCoV-Spike
ACE2 proteins of different species
We first collected 1056 ACE2 proteins from NCBI database, where the sequences without the basic information of species. The human ACE2 protein (np_0013583411.1) [23] was selected as template for ‘full-length’ sequence alignment, and the ACE2 protein sequences of all species were conducted for clustering by using the software BLAST [24]. Finally, we obtained 173 species with the identity criterion >60% for further analysis. We constructed a phylogenetic tree for all the 173 species including human being by using ACE2 proteins, as shown in SI Figure 1. Then, we selected 18 species (chimpanzee, monkey, totoro, rabbit, tiger, lion, cat, cynomys, dog, rat, sheep, whale, pig, cow, deer, dolphin, duck and chicken) keeping close contacts with human beings for detailed analysis. As bat [8,9,10], manis [11] and snake [25] were once considered to be the original host of coronavirus to human beings, thus we also take these 3 species for further analysis.
Figure 1.
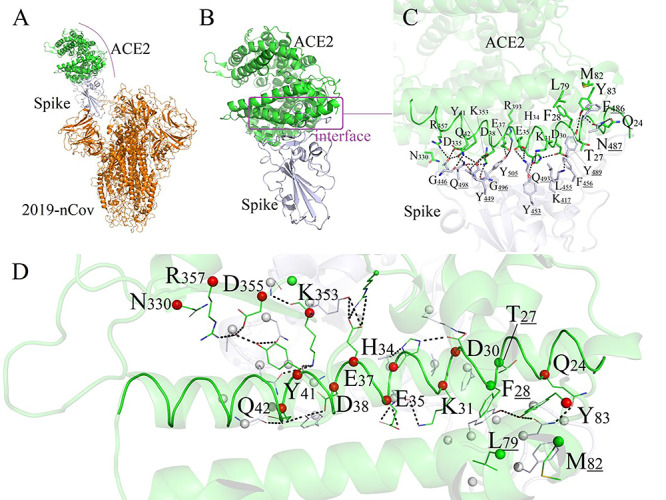
Binding mode between human ACE2 and 2019-nCoV-Spike. (A) Conformation of 2019-nCoV/ACE2 complex. (B/C) Detailed binding modes between ACE2 and 2019-nCoV Spike region. (D) 18 residues of ACE2 forming direct interactions with 2019-nCoV. The 14 key polar residues of human ACE2 were shown as red spheres. The ACE2 protein: green, labeled as black; 2019-nCoV: orange; 2019-nCoV-Spike region: white.
Binding modes between human ACE2 and 2019-nCoV-Spike
We next investigated how human ACE2 proteins bind the novel coronavirus and analyzed the important residues of ACE2 which form the binding interface of complex ACE2/2019-nCoV-Spike [26,27]. The crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 has been resolved [28]. Here, we obtained it from the RCSB PDB.databank and analyzed the binding mode between human ACE2 and 2019-nCoV-Spike (as shown in Figure 1A). The protein–protein binding interface was clear. A total number of 18 amino acids on ACE2 form good interface with 2019-nCoV-Spike, among which 14 amino acids contributed the major polar contacts, and 4 residues participate in the formation of hydrophobic interactions between two proteins, as shown in Figure 1B/C/D.
As shown in Figure 1C, 14 polar amino acids (Q24, D30, K31, H34, E35, E37, D38, Y41, Q42, Y83, N330, K353, D355 and R357) on human ACE2 protein could form intensive hydrogen-bond networks with 12 amino acids (K417, G446, Y449, Y453, N487, Y489, Q493, G496, Q498, T500, N501 and Y505) of the 2019-nCoV-Spike protein. Among these polar interactions, the typical salt-bridging interaction between D30 and K417 play significant roles in maintaining the protein–protein stability. The 14 key polar residues of human ACE2 are shown as red spheres in Figure 1D. In the following section, we will investigate and analyze these 14 key polar residues in 173 species to find whether residues are conserved in different species and the way that they affect the binding modes with the novel coronavirus.
Key polar residues in 173 species
We collected 18 residues on ACE2 from all the 173 species which form the interactions with 2019-nCoV-Spike. By sequence alignment and statistical analysis, we found out of the 14 polar residues, 8 are highly conserved, as shown in Figure 2. These 8 highly conserved polar residues have a big chance to ensure the basic binding affinity with the novel coronavirus. There are 6 polar residues (positions at human ACE2: 24, 30, 31, 34, 35 and 38) which might significantly affect the binding free energies of ACE2/2019-nCoV-Spike in different species. Sequence alignment provides information about the primary structural similarity of the ACE2 proteins in different species, however, this is not indicative by itself of their functional similarity. As for positions 30 and 38, Asp (D) and Glu (E) have high frequency in all 173 species. Their identical physicochemical properties allow them to serve the same biological role in binding modes. In order to check the influences of 14 polar residues, 21 species keeping close contacts with human beings were selected for subsequent binding energies and interacting modes analysis.
Figure 2.
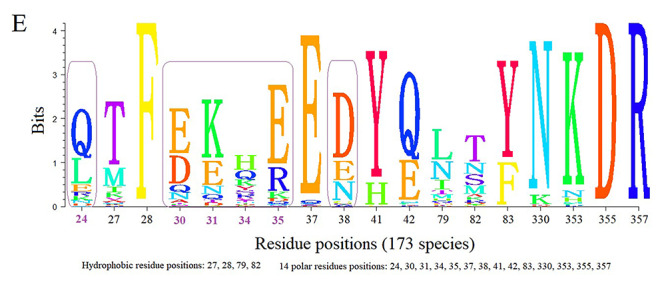
Sequence alignment result of 18 key residues of ACE2 protein in 173 different species which form the interactions with 2019-nCoV-Spike.
Homology modeling of ACE2 proteins of different species
Next, the ACE2 proteins for other species were constructed by Homology Modeling method. The homology modeling of each ACE2/2019-nCoV-Spike complex was conducted by the software Modeler [29], and the modeling structures were shown in Figure 3A.
Figure 3.
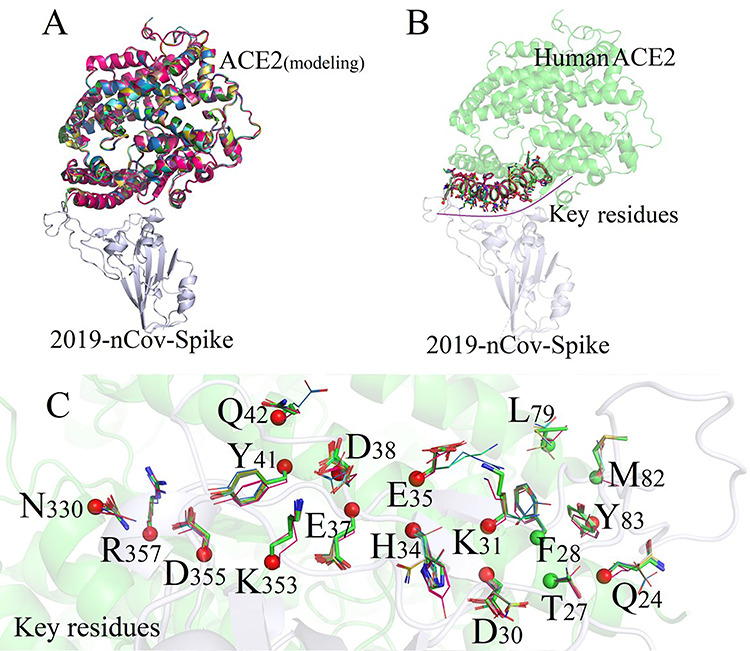
Superimposed structures of ACE2 and key residues between Human and different species’ modeling structures. (A) Homology modeling of 21 species; (B) Residues of ACE2 forming direct interactions to 2019-nCoV-Spike; (C) 18 residues of ACE2 forming direct interactions with 2019-nCoV (The key 14 polar residues were colored by red, and unpolar residues were colored by green).
Currently, only the crystal structure of human ACE2 protein (PDB ID: 6m0j) [28] was published on the RSCB PDB database, and the sequence similarities of ACE2 proteins for all 173 species were higher than 60% compared with that of human (as shown in Figure 3), which completely meets the modeling criterion of >30% [30] sequence similarity for homologous modeling. In this project, the crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 was adopted as a single template to obtain structures for all other 172 species.
Based on the basic principle of homologous modeling, the more similar the sequence between the modeling template and the target proteins are, the closer the structure would be. Therefore, the main framework between modeling structures and the template protein were completely consistent, and there were no significant structural changes among all ACE2 protein domains for all species, as shown in Figure 3A. Subsequently, each modeling structure was superposed on the template chain by using PyMol [31] to obtain the complex structures of ACE2/2019-nCoV-Spike, and the modeling structures were shown in Figure 3B.
The key residues of ACE2 protein for 22 species were shown in Figure 3C and Table 1. It can be seen that the sequences of 14 key polar amino acids forming the binding interface from different species were quite different, especially for duck, chicken and snake, which are <56% similar with that of human beings. We also discovered that the 5 key point residues (position: 24, 30, 31, 34 and 35) present highly unconserved properties which might have decisive effects on the binding free energies of ACE2/2019-nCoV-Spike in different species. The binding free energies of ACE/2019-nCoV-Spike complex for 22 species were listed in Table 2. We next calculate the binding free energies and check the stability of each ACE2/2019-nCoV-Spike complex.
Table 1.
Sequence alignment of 14 key polar residues in 22 different species
| Species | Scientific name | Key polar residues of ACE2 in different species | Sequence similarity | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 | 27 | 28 | 30 | 31 | 34 | 35 | 37 | 38 | 41 | 42 | 79 | 82 | 83 | 330 | 353 | 355 | 357 | Full- | Key polar- | ||
| Rat | Mus musculus | Q | E | F | K | Q | A | R | D | D | Y | A | L | M | Y | N | K | D | R | 84.25% | 57.14% |
| Chicken | Gallus gallus | Q | T | F | A | E | V | R | E | D | Y | E | N | R | F | N | K | D | R | 70.25% | 57.14% |
| Snake | Thamnophis elegans | Q | E | F | K | Q | A | R | D | D | Y | A | L | M | Y | N | K | D | R | 64.13% | 57.14% |
| Duck | Anas platyrhynchos | Q | M | F | A | K | V | R | E | D | Y | E | N | N | F | N | K | D | R | 70.59% | 64.28% |
| Totoro | Chinchilla lanigera | Q | T | F | D | N | E | K | E | D | Y | Q | L | M | Y | N | K | D | R | 87.58% | 78.57% |
| Cat | Felis catus | L | T | F | E | K | H | E | E | E | Y | Q | L | T | Y | N | K | D | R | 86.10% | 78.57% |
| Manis | Manis javanica | E | T | F | E | K | S | E | E | D | Y | Q | I | N | Y | N | K | D | R | 85.40% | 78.57% |
| Rabbit | Oryctolagus cuniculus | L | T | F | E | K | Q | E | E | D | Y | Q | L | T | Y | N | K | D | R | 87.10% | 78.57% |
| Lion | Puma concolor | L | T | F | E | K | H | E | E | E | Y | Q | L | T | Y | N | K | D | R | 86.43% | 78.57% |
| Tiger | Panthera tigris altaica | L | T | F | E | K | H | E | E | E | Y | Q | L | M | Y | N | K | D | R | 86.43% | 78.57% |
| Sheep | Ovis aries | Q | T | F | E | K | H | E | E | D | Y | Q | M | T | Y | N | K | D | R | 82.89% | 92.85% |
| Whale | Physeter catodon | Q | T | F | Q | K | H | E | E | D | Y | Q | T | T | Y | N | K | D | R | 82.89% | 92.85% |
| Deer | Odocoileus virginianus | Q | T | F | E | K | S | E | E | D | Y | Q | M | T | Y | N | K | D | R | 82.38% | 94.44% |
| Chimpanzee | Pan paniscus | Q | T | F | D | K | H | E | E | D | Y | Q | L | M | Y | N | K | D | R | 99.50% | 100% |
| Human | Homo sapiens | Q | T | F | D | K | H | E | E | D | Y | Q | L | M | Y | N | K | D | R | 100% | 100% |
| Cow | Bos taurus | Q | T | F | E | K | H | E | E | D | Y | Q | L | M | Y | N | K | D | R | 82.72% | 92.85% |
| Dolphin | Phocoena sinus | Q | T | F | Q | K | H | E | E | D | Y | Q | I | M | Y | N | K | D | R | 81.21% | 92.85% |
| Monkey | Macaca mulatta | Q | T | F | D | K | H | E | E | D | Y | Q | L | M | Y | N | K | D | R | 96.82% | 100% |
| Cynomys | Marmota | L | T | F | D | K | Q | E | E | D | Y | Q | L | M | Y | N | K | D | R | 85.93% | 85.71% |
| Pig | Sus scrofa domesticus | L | T | F | E | K | L | E | E | D | Y | Q | I | T | Y | N | K | D | R | 82.75% | 78.57% |
| Dog | Canis lupus familiaris | L | T | F | E | K | Y | E | E | E | Y | Q | L | T | Y | N | K | D | R | 84.68% | 71.42% |
| Bat | Phyllostomus discolor | D | K | F | E | N | N | E | E | E | Y | Q | L | N | Y | N | K | D | R | 82.03% | 64.28% |
Nonpolar residues: 27, 28, 79 and 82; 14 polar residues: 24, 30, 31, 34, 35, 37, 38, 41, 42, 83, 330, 353, 355 and 357.
Table 2.
Binding free energies between each species’s ACE2 protein and 2019-nCoV-Spike region
| Species | Scientific Name | Full-sequence similarity | Key-residue similarity | Items of binding free energy (Kcal/mol) | ||||
|---|---|---|---|---|---|---|---|---|
| Complex | Receptor | Ligand | TOTAL | |||||
| A | Rat | Mus musculus | 84.25% | 57.14% | −73022.52 | −56358.20 | −16633.51 | −30.80 |
| Chicken | Gallus gallus | 70.25% | 57.14% | −72535.74 | −55811.99 | −16690.90 | −32.84 | |
| Snake | Thamnophis elegans | 64.13% | 57.14% | −73319.10 | −56639.95 | −16646.14 | −33.01 | |
| Duck | Anas platyrhynchos | 70.59% | 64.28% | −72657.17 | −56027.19 | −16596.10 | −33.87 | |
| Totoro | Chinchilla lanigera | 87.58% | 78.57% | −73469.24 | −56820.96 | −16608.81 | −39.46 | |
| B | Cat | Felis catus | 86.10% | 78.57% | −73304.82 | −56642.13 | −16614.76 | −47.92 |
| Manis | Manis javanica | 85.40% | 78.57% | −73573.68 | −56895.04 | −16627.97 | −50.66 | |
| Rabbit | Oryctolagus cuniculus | 87.10% | 78.57% | −73493.65 | −56769.76 | −16672.95 | −50.94 | |
| Lion | Puma concolor | 86.43% | 78.57% | −73359.01 | −56662.09 | −16645.46 | −51.45 | |
| Tiger | Panthera tigris altaica | 86.43% | 78.57% | −73355.00 | −56701.68 | −16704.87 | −51.55 | |
| C | Sheep | Ovis aries | 82.89% | 92.85% | −73410.67 | −56697.10 | −16660.82 | −52.74 |
| Whale | Physeter catodon | 82.89% | 92.85% | −73014.88 | −56345.43 | −16615.42 | −54.02 | |
| Deer | Odocoileus virginianus | 82.38% | 94.44% | −73856.79 | −57177.69 | −16624.24 | −54.86 | |
| Chimpanzee | Pan troglodytes | 99.50% | 100% | −73736.47 | −57025.61 | −16655.79 | −55.06 | |
| Human | Homo sapiens | 100% | 100% | −73798.62 | −57139.98 | −16603.55 | −55.07 | |
| Cow | Bos taurus | 82.72% | 92.85% | −73529.96 | −56837.04 | −16637.77 | −55.14 | |
| Dolphin | Phocoena sinus | 81.21% | 92.85% | −72836.93 | −56074.67 | −16707.09 | −55.16 | |
| Monkey | Macaca mulatta | 96.82% | 100% | −73760.68 | −57117.22 | −16586.16 | −57.30 | |
| D | Cynomys | Marmota | 85.93% | 85.71% | −73364.50 | −56661.48 | −16644.90 | −58.10 |
| Pig | Sus scrofa domesticus | 82.75% | 78.57% | −73223.45 | −56547.54 | −16616.89 | −59.01 | |
| Dog | Canis lupus familiaris | 84.68% | 71.42% | −73377.09 | −56655.94 | −16659.48 | −61.66 | |
| Bat | Phyllostomus discolor | 82.03% | 64.28% | −73117.67 | −56389.30 | −16665.12 | −63.25 | |
22 species were divided into four categories according to the binding energy values: Class A: Rat, Snake, Chicken, Duck and Totoro; Class B: Cat, Deer, Manis, Rabbit, Lion and Tiger; Class C: Sheep, Whale, Deer, Chimpanzee, Human, Cow, Dolphin and Monkey; Class D: Cynomys, Pig, Dog and Bat. Binding free energies present linear cor-relationship with the key 14 polar amino acids.
Binding free energy calculation for 22 species
To investigate how the 14 polar residues affect the binding modes of ACE2 to the 2019-nCoV-Spike, we first used the Root Mean Square Deviation (RMSD) [32] of heavy atoms for 22 species to check the stability of each ACE2/2019-nCoV-Spike complex. As shown in SI Figure 2, the calculated maximum RMSD fluctuation value for each complex is under 1.8 Å and they all reach equilibrium states at the 3 ns. All these data indicate that the 2019-nCoV-Spike coronavirus is stable when combines with the human ACE2. We picked out the average structure of each complex system after molecular dynamics equilibrium for subsequent analysis. As shown in SI Figure 2A-F, the RMSD values for 6 species (manis, rat, chicken, duck, snake and dog) present highly fluctuating properties, indicating an unstable binding modes, and there might be one process of ‘Targeting-Miss Targeting’ in the binding of ACE2/2019-nCoV-Spike complex. As shown in SI Figure 2G-J, 4 species (sheep, whale, deer and cow) tend to be relatively stable in the binding process, but there are temporarily conformational swings after the 2019-nCoV-Spike structure bound with the ACE2 protein. It is speculated that the novel coronavirus could infect these species. Species in SI Figure 2K-S show extremely stable states after equilibrium, indicating that ACE2/2019-nCoV-Spike complex structures are relatively stable after novel coronavirus bound with the ACE2 proteins in different species. In general, these species might own better binding capacities with the novel coronavirus.
Figure 4.
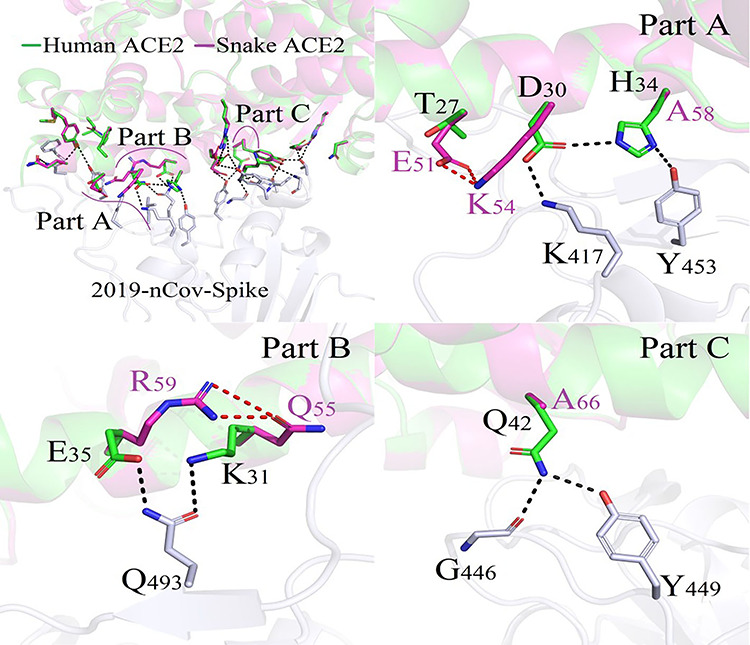
Difference in binding modes of ACE2/2019-nCoV-Spike between Human and Snake.
From the perspective of the full-length sequence similarity, no direct relationship was extracted for the binding affinities between 2019-nCoV-Spike and ACE2 proteins from each species, as shown in Table 2. However, the binding free energies were found to present the linear correlation with those 14 key polar residues that formed the binding interface. It is obvious to extract basic rules as follows:
(1) The binding energies for all species, with >90% sequence similarities of key-residues compared with the human beings, fluctuating slightly from −52 kcal/mol to −58 kcal/mol;
(2) When the sequence similarity of key amino acids fluctuated sharply, the corresponding binding affinities also changed greatly.
From the analysis above, we can see that the 14 key polar residues could determine the basic binding affinities, and the 5 polar unconserved residues (position: 24, 30, 31, 34 and 35) might even own decisive effects on the binding modes between 2019-nCoV-Spike and ACE2 in different species.
The binding free energy between human ACE2 protein and the Spike region on coronavirus 2019-nCoV was −55.07 kcal/mol, which was consistent with the data reported in [33]. However, the binding free energies of ACE2/2019-nCoV-Spike complex for other 21 species were fluctuated from −66.26 kcal/mol to −32.84 kcal/mol, indicating a large variation among different species.
The most significant improvement of binding affinity was discovered in 3 species including pig, dog and bat. The bat, which possesses the highest binding ability −63.26 kcal/mol, was suspected as the first case of novel coronavirus infection source [8,9,10]. It is worth noting that one highly increased binding energy of −61.66 kcal/mol was assigned to the species dog which was reported as the first lethal case infected with 2019-nCoV virus [12,13]. In addition, we also show definite evidence that the 2019-nCoV-Spike binds ACE2 proteins of 8 species (chimpanzee, marmot, pig, cow, rabbit, cynomys, deer and sheep) with similar binding affinities, fluctuating from −58 kcal/mol to −52 kcal/mol, compared with that of human beings. All these species are close contacts of human beings. In addition, the manis has also been considered as the origin of 2019-nCoV coronavirus [11], and the calculated binding energy of its ACE2/2019-nCoV-Spike was −50.66 kcal/mol.
Animals with the lowest binding energies <−40.00 kcal/mol were observed in four species: totoro, duck, chicken and snake. In particular, snake was also considered to be the original source of novel coronavirus [25], but the corresponding binding energy was the lowest −33.01 kcal/mol among 22 species.
Unfortunately, we found that mouse (Mus musculus) still owns the binding capacity to 2019-nCoV-Spike with weak binding energy of −30.80 kcal/mol. The mouse with absolutely strong survival abilities are widely distributed in the world. If mice were identified as the virus hosts and presented ‘Human-Mouse’ transmitting and cross-infecting abilities, this might bring disasters to our human species which have already happened to the species like deer and cat.
In summary, all of the information indicates the basic binding capacities for all 22 species to the 2019-nCoV-Spike virus. If ‘Parasite–Host’ relationships were found within these species to the novel coronavirus, the effectively cross-infections might bring fatal threats to our human beings and the global ecosystem.
Difference analysis of binding modes
In order to clarify the binding modes between 2019-nCoV-Spike and ACE2 proteins in different species, we made a comparative analysis of their binding patterns with that of human beings. The 21 species and human beings were divided into four categories according to the binding energy values: Class A (Snake, Chicken, Duck and Totoro), Class B (Manis and Rabbit), Class C (Sheep, Whale, Chimpanzee, Cow, Dolphin and Monkey) and Class D (Cynomys, Pig, Dog and Bat). Residues from ACE2 proteins forming the binding interface presented the high-frequency variation properties in these species.
In all of the 22 species, the snake owns the lowest binding capacity of ACE2 protein to coronavirus 2019-nCoV-Spike which is −33.01 kcal/mol. As shown in Figure 4, compared with that of human beings, 3 regions (Part A, Part B and Part C) with large structural differences in the binding mode were extracted. In these three regions, one complicated and compact polar interaction network was formed between human ACE2 protein and the coronavirus Spike protein. However, these polar contacts were disappeared in species snake, and this was the main reason for the decline of the snake ACE2/2019-nCoV-Spike complex binding capacity.
Manis possessed relatively weaker binding capacity −50.67 kcal/mol than that of human beings −55.08 kcal/mol. Its binding mode of ACE2 and 2019-nCoV-Spike was shown in Figure 5. The salt-bridging bond formed between the nitrogen atom of H34 and the oxygen atom of carboxylate acid group on D40, attenuated the polar interaction of hydrogen bond D40-K417 between ACE2 and 2019-nCoV-Spike. At the same time, one weak hydrogen bond interaction between the hydroxyl group of Y453 from Spike to S34 on ACE2 protein.
Figure 5.
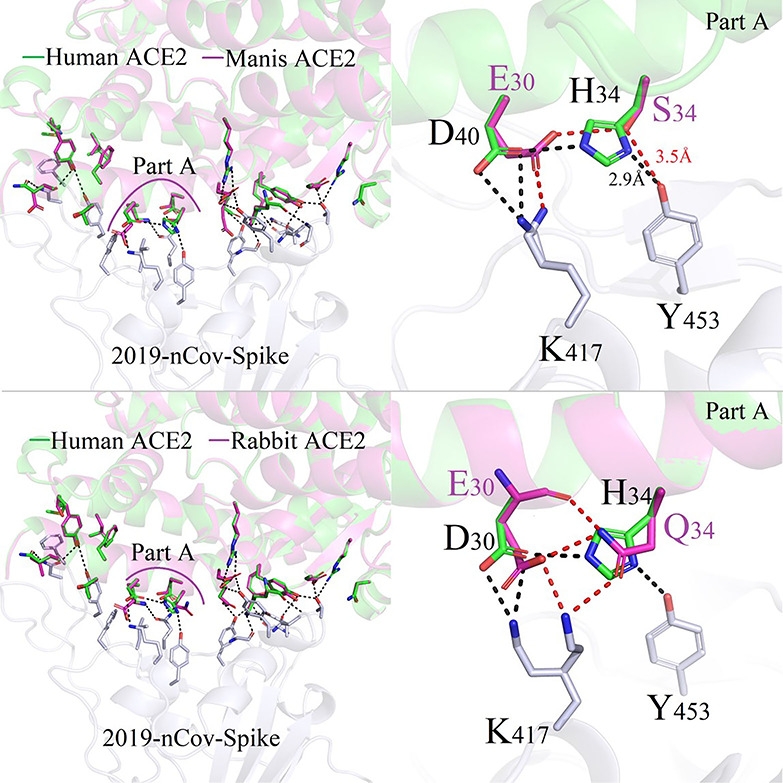
Difference in binding modes of ACE2/2019-nCoV-Spike between Human and Manis/Rabbit species.
Figure 6 plots the conformation differences at the same position D30/E30/Q30 in human beings, sheep, wale and Lion. From Figure 6, we can see that sheep and wale have similar binding interaction modes between 2019-nCoV-Spike and the corresponding ACE2. Binding free energies for sheep and whale decrease slightly in sheep of ACE2/2019-nCoV-Spike was −52.74 kcal/mol; this value is consistent with the calculated values of −54.02 kcal/mol and − 55.07 kcal/mol for whale and human.
Figure 6.
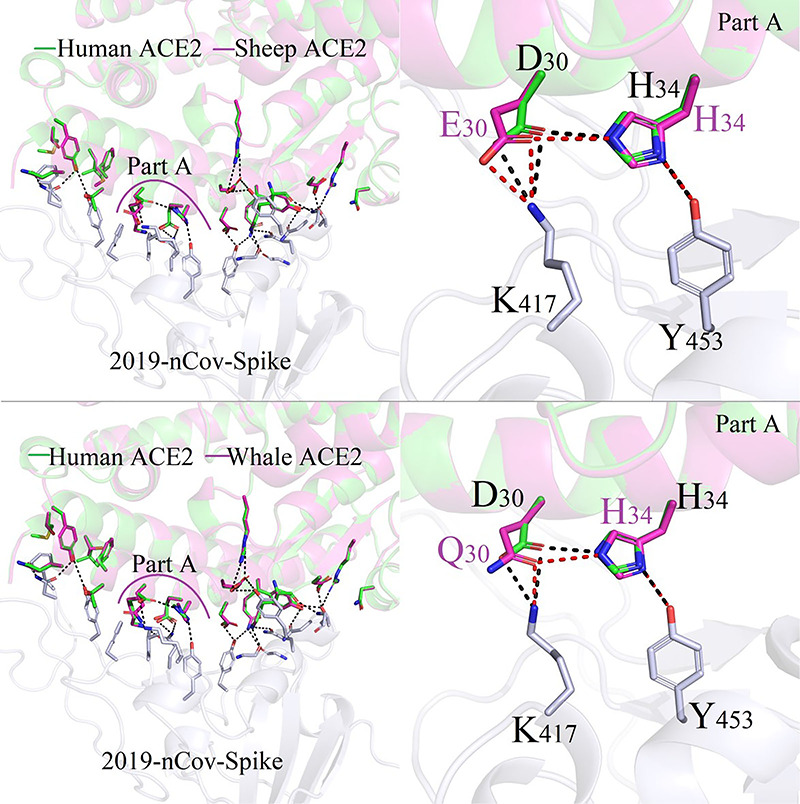
Difference in binding modes of ACE2/2019-nCoV-Spike between Human and Sheep/Whale species.
The bats are unanimously considered to be the main infecting source of the coronavirus [8,9,10], and a maximum energy variation (bat: −63.25 kcal/mol, human: −55.07 kcal/mol) occurred with the ordering of different residues in ACE2/2019-nCoV-Spike complexes. The structural data indicated that atoms of the functional group amin -NH3 from residue K27 on bat ACE2 make new polar contacts with residue Y473 in Spike region, as shown in Figure 7. The protonated nitrogen formed directly salt-bridges to the carboxylate between the E30 (bat ACE2) and K417 (2019-nCoV-Spike) in a manner similar to that reported in most species. Furthermore, regional interactions were significantly improved by varying the amino acid D38 to E38, as one set of more complicated polar interactions including Q756, Q42, Y449, D/E38, G496 and K350 were detected.
Figure 7.
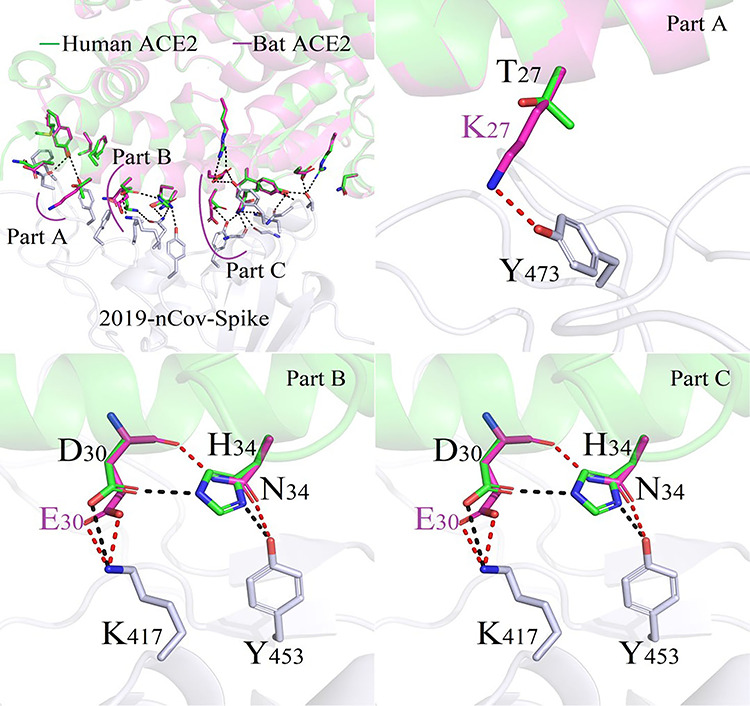
Difference in binding modes of ACE2/2019-nCoV-Spike between Human and Bat species.
As shown in Figure 7, the new stronger salt-bridge of E30-K417 play an essential role for improving the binding ability between pig ACE2 protein and 2019-nCoV-Spike.
In order to investigate the basic principles of ACE2/2019-nCoV-Spike binding modes, qualitative statistics of the hydrogen bonds and salt bridges formed between these 14 polar amino acids and 2019-nCoV-Spike were summarized in Table 3. As shown in Table 3, it was obvious that all ACE2 proteins from 22 different species possessed the binding capabilities with the Spike region on novel coronavirus in terms of binding free energy for each ACE2/2019-nCoV-Spike complex. Compared with the human beings, the ACE2 protein from 21 species presented different ranges of binding free energies with each other. 14 amino acids formed one good polar interacting network to maintain the stabilities of ACE2/2019-nCoV-Spike interactions. Among them, 8 polar residues at the same position with that of human ACE2 protein are highly conserved, which ensured each species’ basic binding characteristics with the coronavirus. 5 of the other 6 polar residues (human ACE2 positions: Q24, D30, K31, H34 and E35), presenting highly unconserved properties, could significantly affect the binding free energies of ACE2/2019-nCoV-Spike in different species.
Table 3.
Polar interaction modes variations of ACE2/2019-nCoV-Spike among different species
| Polar contacts | Binding modes variation analysis (Specied/Energy: kcal/mol) | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residues | ACE2 … Spike | Rat | Chicken | Snake | Duck | Totoro | Cat | Manis | Rabbit | Lion | Tiger | Sheep | Whale | Deer | Chimpanzee | Human | Cow | Dolphin | Monkey | Cynomys | Pig | Dog | Bat |
| Q24 | Q24-OE1 … ND2-N487 | - | - | - | - | - | × | - | × | × | × | - | - | - | - | - | - | - | - | × | × | × | - |
| D30 | D30-OD1 … NZ-K417 | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ↑ | ↑ |
| D30-OD2 … NZ-K417 | × | × | × | × | - | - | × | ↓ | ↓ | ↓ | ↓ | - | - | - | - | - | ↓ | - | ↑ | ↑ | ↑ | ↑ | |
| K31 | K31-NZ … OE1-Q493 | × | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| H34 | H34-ND1 … OH-Y453 | - | × | - | × | × | - | ↓ | × | - | - | - | - | - | - | - | - | - | - | - | - | ↑ | - |
| E35 | E35-OE1 … NE2-Q493 | × | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E35-OE2 … NE2-Q493 | × | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| E37 | E37-OE1 … OH-Y505 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E37-OE2 … OH-Y505 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| D38 | D38-OD2 … OH-Y449 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| D38-OD1 … OH-Y449 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Y41 | Y41-OH … OG1-T500 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Y41-OH … OD1-N501 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Q42 | Q42-NE2 … O-G446 | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Q42-NE2 … OH-Y449 | × | × | × | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Y83 | Y83-OH … OD1-N487 | - | × | - | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Y83-OH … OH-Y489 | - | × | - | × | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| N330 | N330-ND2 … OG1-T500 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K353 | K353-O … N-G502 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| K353-NZ … OE1-Q498 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| K353-NZ … O-G496 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| D355 | D355-OD2 … OG1-T500 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| R357 | R357-NH1 … OG1-T500 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Binding energy(kcal/mol) | −30.80 | −33.01 | −32.84 | −33.87 | −39.46 | −47.92 | −50.66 | −50.94 | −51.45 | −51.55 | −52.74 | −54.02 | −54.68 | −55.06 | −55.07 | −55.14 | −55.16 | −57.30 | −58.10 | −59.01 | −61.66 | −63.25 | |
The polar interactions of residues at the same position in different specie’s ACE2 protein were listed.
No change: ‘-’; Enhance: ‘↑’; Weaken: ‘↓’; disappear: ‘×’.
Discussion
In this paper, we collected 1056 ACE2 protein sequences from NCBI database and obtained 173 species which have more than 60% sequence identity compared with that of human beings by sequence alignment of their ACE2 proteins. We analyzed 14 key polar residues for these 173 species and selected 21 species keeping close contacts with human beings to investigate the differences of their interacting modes and binding affinities with that of human beings. Out of the 18 amino acids on ACE2 proteins which form the binding interface with the Spike protein on the novel coronavirus, only 4 residues formed hydrophobic interactions with 2019-nCoV-Spike protein, and 14 amino acids belonging to the polar interacting (hydrogen bond, salt bridge, etc.) modes that played absolutely major roles in maintaining the stabilities of ACE2/2019-nCoV-Spike interactions. We found that 8 polar amino acid sites at the same position with that of human ACE2 protein are highly conserved, which ensured each species’ basic binding characteristics with the coronavirus. 5 of the other 6 polar amino acids (human ACE2 positions: Q24, D30, K31, H34 and E35) could significantly affect the binding free energies of ACE2/2019-nCoV-Spike in different species.
Almost all the ACE2 proteins from the 21 different species possessed the binding capabilities with the Spike region on novel coronavirus in terms of binding free energy for each ACE2/2019-nCoV-Spike complex. Compared with the human beings, the ACE2 protein from mouse, totoro, chicken, duck and snake possessed the weak binding affinities with the 2019-nCoV-Spike region; lion, tiger, manis, deer, cat and rabbit showed slightly decreasing binding capabilities with the novel coronavirus; cow, cynomys, chimpanzee, monkey, sheep, dolphin and whale presented almost the same binding free energies with each other; bat, pig and dog showed the most significant improvements in terms of binding abilities.
It could be inferred that the novel coronavirus might possess infective abilities to many species. In addition, the complicated situations of ‘Animal–Animal’ and ‘Animal-Human’ cross-species infections might have happened among different species. Up till now, we have finished calculating the binding free energies for 130 species and the results for the rest 41 species would be updated in our website: http://bioinformatics.csu.edu.cn/species/.The corresponding binding affinities for the calculated 130 different species was −75.48 ~ −10.13 kcal/mol, as shown in Figure 8. Among the calculated 130 species, 75 species possess relatively strong binding affinities (<−50 kcal/mol) compared with that of human beings (−55.07 kcal/mol). It is astonishing to find that the binding abilities of 2019-nCoV-Sike to the ACE2 proteins for 44 species were significantly lower than −55.0 kcal/mol. All other species owning sequence similarity <60% will be considered for calculating in next steps. We hope this research could provide significant help for the future epidemic detection, vaccine and drug development research, and the global eco-system protections.
Figure 8.
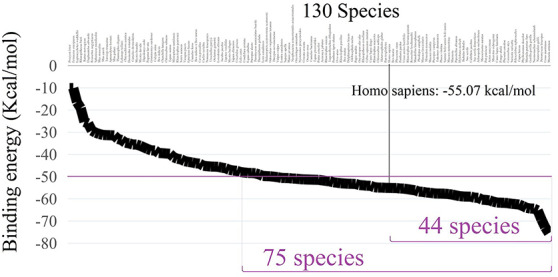
The corresponding binding free energies for 130 species.
Methods
Phylogenetic tree construction
The program of MEGA7 [34] was used to align the FASTA files of 173 species, and the alignment results were export to Meg formats. Then the Meg format files of ACE2 sequences were imported into MEGA7 again to build the evolutionary tree. Finally, the evolutionary tree for all species was processed with Evolview V2 program [35] to add markers and corresponding coloring subtrees.
Homology modeling
Homology modeling process of ACE2 protein for each species was conducted by the software Modeller 9.14. The crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 extracted from the Protein Data Bank (PDB ID: 6m0j) [28] was selected as the single template for modeling. The predicted structures of ACE2 protein for each species were generated and saved in the PDB format and sorted according to scores calculated from Discrete Optimized Protein Energy (DOPE). The best model was selected with regard to the DOPE score9
Molecular dynamics simulations
In order to check the stability of ACE2/2019-nCoV-Spike complex, the protein structure of each specie was employed for >6 ns MD simulations. The molecular dynamics simulations were carried out by AMBER software (version 16) [36], using AMBER ff99sb force field for each complex. The complex was solvated in a cubic periodic box of explicit TIP3P water model that extended a minimum 10 Å distance from the box surface to any atom of the ACE2/2019-nCoV-Spike complex. To eliminate possible bumps, all heavy atoms were position restrained with a strong potential of the form k (Δx)2 with a force constant k = 1500 kcal/mol−1 Å−2. The constant temperature was selected at 298 K with the NPT ensemble. Finally, based on the final >6 ns MDs trajectory, at least 3000 snapshots were extracted from the equilibrium trajectory for the final average structure of each species’ complex. Based on the 6n molecular dynamics simulations trajectories, the binding free energies of ACE2/2019-nCoV-Spike were computed for each snapshot and averaged using the MM-PBSA approach implemented as script (MMPBSA.py) in AMBER software.
Key points
The new coronavirus might parasitize in other species which might result in the cross-species infections. Comparing with the human beings, 44 species present significant improvements in binding affinities.
In this paper, we found 14 polar residues forming the binding interface of ACE2/2019-nCoV-Spike complex which play an important role in maintaining protein–protein stability. Among them, 8 polar residues at the same positions with that of human ACE2 are highly conserved, which ensure its basic binding affinity with the novel coronavirus.
Five unconserved polar residues at the binding interface of ACE2/2019-nCoV-Spike complex are proved to have an effect on the binding patterns among species.
Supplementary Material
Funding
This study was funded by the National Natural Science Foundation of China (No. 61832019).
Senbiao Fang received the B.S. degree in Chemistry from Xiangtan University, Xiangtan, China, in 2007, and received the M.S. degree in computational biology from Lanzhou University, Lanzhou, China, in 2014. He is currently working toward the PhD degree in the School of Computer Science and Engineering, Central South University, China. His research interests include bioinformatics and system biology.
Ruoqian Zheng received the B.S. degree in network engineering from Xiangtan University, Xiangtan, China, in 2019. Currently, she is working toward the M.S. degree in computer science in the Central South University, Changsha, China. Her current research interest is structure biology.
Chuqi Lei received the B.S. degree in computer science from Central South University, China, in 2020. Currently, she is working toward the Ph.D. degree in computer science in the Central South University, Changsha, China. Her current research interest is proteomics.
Jianxin Wang received the B.S. and M.S. degrees in computer engineering from Central South University, Changsha, China, in 1992 and 1996, respectively, and the PhD degree in computer science from Central South University, Changsha, China, in 2001. He is the dean and a professor in School of Computer Science and Engineering, Central South University, Changsha, Hunan, P.R. China. His current research interests include algorithm analysis and optimization, parameterized algorithm, bioinformatics and computer network.
Ruiqing Zheng received the B.S. degree and the M.S. in computer science from Central South University, Changsha, China, in 2013, 2016, respectively. He is currently working toward the PhD degree in the School of Computer Science and Engineering, Central South University, China. His research interests include bioinformatics and system biology.
Min Li received the B.S. degree in communication engineering and the M.S. and Ph.D. degrees in computer science from Central South University, Changsha, China, in 2001, 2004, and 2008, respectively. She is currently a professor and vice dean at the School of Computer Science and Engineering, Central South University. Her main research interests include bioinformatics and system biology.
Contributor Information
Senbiao Fang, Xiangtan University, Xiangtan, China.
Ruoqian Zheng, Xiangtan University, Xiangtan, China.
Chuqi Lei, Central South University, China.
Jianxin Wang, Central South University, Changsha, China.
Ruiqing Zheng, Central South University, Changsha, China.
Min Li, Central South University, Changsha, China.
Data availability
The datasets used in the study are available from the National Center for Biotechnology Information (NCBI) database under the accession number: NP_001358344.1.
References
- 1. Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV[J]. Nat Commun 2020;11(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorbalenya AE. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2[J]. Nature Microbiology 2020;5:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farag E, Sikkema RS, Mohamedani AA, et al. MERS-CoV in camels but not camel handlers, Sudan, 2015 and 2017[J]. Emerg Infect Dis 2019;25(12):2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses[J]. Science 2005;310(5748):676–9. [DOI] [PubMed] [Google Scholar]
- 5. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding[J]. The Lancet 2020;395(10224):565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China[J]. Nature 2020;579(7798):265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luan J, Lu Y, Jin X, et al. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection[J]. Biochem Biophys Res Commun 2020;526(1):165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J, Liu S, Fan J, et al. NK-CDS: a creative design system for museum art derivatives[J]. IEEE Access 2020;8:29259–69. [Google Scholar]
- 9. Jaimes JA, Andre NM, Millet JK, et al. Structural modeling of 2019-novel coronavirus (nCoV) spike protein reveals a proteolytically-sensitive activation loop as a distinguishing feature compared to SARS-CoV and related SARS-like coronaviruses[J]. arXiv preprint arXiv 2020;2002:06196. [Google Scholar]
- 10. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature 2020;579(7798):270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lam TTY, Shum MHH, Zhu HC, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins[J]. Nature 2020;583:1–6. [DOI] [PubMed] [Google Scholar]
- 12. Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2[J]. Science 2020;368(6494):1016–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goumenou M, Spandidos DA, Tsatsakis A. Possibility of transmission through dogs being a contributing factor to the extreme Covid-19 outbreak in North Italy[J]. Mol Med Rep 2020;21(6):2293–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coronavirus: Bronx zoo tiger tests positive for Covid-19[N]. The Guardian Press. The Bronx Zoo, New York, USA, 2020, Sarah Caddy. [Google Scholar]
- 15. Stadnytskyi V, Bax CE, et al The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission[J]. PNAS 2020;117(22):11875–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu P, Qi F, Xu Y, et al. Age-related rhesus macaque models of COVID-19[J]. Animal models and experimental medicine 2020;3(1):93–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandrashekar A, Liu JY, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques[J]. Science 2020;369(6505):812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice[J]. Nature 2020;583:830–3. [DOI] [PubMed] [Google Scholar]
- 19. Chan JFW, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility[J]. Clin Infect Dis 2020;325:1093–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lakdawala SS, Menachery VD. The search for a COVID-19 animal model[J]. Science 2020;368(6494):942–3. [DOI] [PubMed] [Google Scholar]
- 21. Seah I, Agrawal R. Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals[J]. Ocul Immunol Inflamm 2020;28(3):391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun J, Zhuang Z, Zheng J, et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment[J]. Cell 2020;182(3):734, e5–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2[J]. Science 2020;367(6485):1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches[J]. Science 1985;227(4693):1435–41. [DOI] [PubMed] [Google Scholar]
- 25. Ji W, Wang W, Zhao X, et al. Cross-species transmission of the newly identified coronavirus 2019-nCoV[J]. J Med Virol 2020;92(4):433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation[J]. Science 2020;367(6483):1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan S, Sun H, Bu X, et al. An evolutionary RGD motif in the spike protein of SARS-CoV-2 may serve as a potential high risk factor for virus infection?[J]. Front. Pharmacol 2020;11:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor[J]. Nature 2020;581:215–20. [DOI] [PubMed] [Google Scholar]
- 29. Webb B, Sali A. Comparative protein structure modeling using MODELLER[J]. Curr Protoc Bioinformatics 2016;54:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Costanzo M, De Giglio MAR, Roviello GN, et al. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr Med Chem 2020;27(27):4536–4541. [DOI] [PubMed] [Google Scholar]
- 31. Zhu K, Day T, Warshaviak D, et al. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction[J]. Proteins: Structure, Function, and Bioinformatics 2014;82(8):1646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coutsias EA, Seok C, Dill KA. Using quaternions to calculate RMSD[J]. J Comput Chem 2004;25(15):1849–57. [DOI] [PubMed] [Google Scholar]
- 33. Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission[J]. Sci China Life Sci 2020;63(3):457–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sudhir KG, et al. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets.[J]. Mol Biol Evol 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zilong H, Huangkai Z, Shenghan G, et al. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees[J]. Nucleic Acids Research 2016;44:236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salomon-Ferrer R, Götz AW, Poole D, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald[J]. Journal of chemical theory and computation 2013;9(9):3878–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the study are available from the National Center for Biotechnology Information (NCBI) database under the accession number: NP_001358344.1.


