Abstract
Objective
This study aims at reviewing novel coronavirus disease (COVID-19) datasets extracted from PubMed Central articles, thus providing quantitative analysis to answer questions related to dataset contents, accessibility and citations.
Methods
We downloaded COVID-19-related full-text articles published until 31 May 2020 from PubMed Central. Dataset URL links mentioned in full-text articles were extracted, and each dataset was manually reviewed to provide information on 10 variables: (1) type of the dataset, (2) geographic region where the data were collected, (3) whether the dataset was immediately downloadable, (4) format of the dataset files, (5) where the dataset was hosted, (6) whether the dataset was updated regularly, (7) the type of license used, (8) whether the metadata were explicitly provided, (9) whether there was a PubMed Central paper describing the dataset and (10) the number of times the dataset was cited by PubMed Central articles. Descriptive statistics about these seven variables were reported for all extracted datasets.
Results
We found that 28.5% of 12 324 COVID-19 full-text articles in PubMed Central provided at least one dataset link. In total, 128 unique dataset links were mentioned in 12 324 COVID-19 full text articles in PubMed Central. Further analysis showed that epidemiological datasets accounted for the largest portion (53.9%) in the dataset collection, and most datasets (84.4%) were available for immediate download. GitHub was the most popular repository for hosting COVID-19 datasets. CSV, XLSX and JSON were the most popular data formats. Additionally, citation patterns of COVID-19 datasets varied depending on specific datasets.
Conclusion
PubMed Central articles are an important source of COVID-19 datasets, but there is significant heterogeneity in the way these datasets are mentioned, shared, updated and cited.
Keywords: COVID-19, data sharing, review
Introduction
The novel coronavirus disease (COVID-19) outbreak was first reported in Wuhan, China, on 31 December 2019. On 11 March 2020, World Health Organization officially declared COVID-19 a pandemic, marking the recognition of a global crisis [1]. To fight the COVID-19 pandemic, researchers worldwide have quickly investigated different aspects of this disease and reported novel scientific findings, on a daily basis. According to LitCovid [2], a curated literature hub for tracking COVID-19 publication, 34 890 new articles (as the date of 25 July 2020) have been published in the past seven months. Along with published articles, massive and heterogeneous datasets have been created, ranging from testing and case statistics at various locations (medical centers, cities, counties, states, countries), clinical data from studies (e.g., ‘omics, imaging, assays, questionnaires) or from electronic health records, surveys for patient-reported outcomes, administrative data [e.g., ventilators, hospitalizations, intensive care unit (ICU) beds], vital statistics (e.g., obituaries, death certificates), as well as sociodemographic, environmental, economic, individual mobility and transportation data.
Efficient data sharing of biomedical data is an important component in the development of a successful data-driven research on COVID-19 [3]. Researchers reconstructed the early evolutionary paths of COVID-19 by genetic network analysis, for example using existing data of virus genomes collected across the world, providing insights into virus transmission patterns [4]. Nevertheless, it is challenging for researchers to find and identify reliable datasets for novel scientific discoveries, given the large volume and sometimes contradictory information (e.g. non-peer-reviewed sources) about available datasets. Principles such as FAIR (Findable, Accessible, Interoperable and Reusable) [5] and TRUST (Transparency, Responsibility, User focus, Sustainability and Technology) [6] have been proposed for sharing digital data and digital repositories, with applications to COVID-19 datasets as well (e.g. the Virus Outbreak Data Network) [7].
Here, we propose to conduct a systematic review on COVID-19 datasets that are associated with published literature. Our study aims at identifying a comprehensive list of available COVID-19 datasets across domains and at providing insights on how researchers share datasets as they publish COVID-19 research articles. Additionally, we also assess the accessibility, sustainability and impact of published datasets. More specifically, we attempt to answer the following research questions about COVID-19 datasets that are associated with publications:
Q1. Contents: What types of data are published to support different studies and where are those data collected from?
Q2. Accessibility: How can users access datasets and where are the data hosted?
Q3. Citation: How are datasets cited by others and what are top high-impact datasets, by citation count?Our ultimate goal is to promote data sharing and data reuse through careful analyses of current practice by researchers. Through a systematic review, we provide researchers with a comprehensive list of reliable datasets that are available to the public. Additionally, we provide insights about data sharing strategies to aid those who plan to develop and publish new COVID-19 datasets.
Methods
COVID-19 publication collection
To identify and collect COVID-19-related articles, we leveraged LitCovid [2], a newly established literature database for tracking the latest scientific articles about COVID-19, developed by National Library of Medicine in the United States. LitCovid provides essential bibliographic information such as PubMed ID, title, abstract and journal of publications related to COVID-19. In this review, we included all LitCovid articles published before 31 May 2020, resulting in 18 332 articles. As the recognition of associated datasets requires access to full-text articles, we further limited articles to those with full text available in PubMed Central (PMC), which is one of the most significant open access literature repositories of full-text biomedical articles. We then removed errata notes of 16 articles. This further reduced the number of articles to 12 324, from which we carried out our dataset collection process.
COVID-19 dataset collection
We manually reviewed 100 PMC full-text articles and identified the following patterns for mentioning datasets:
(1) Dataset information is available in the Data Availability Statement section provided by PMC, allowing the authors to disclose information about data availability and access, which often contains URL links to data sources, or
(2) When Data Availability Statement section was missing, datasets could have been mentioned in the full text as (a) external URL links to the data sources, (b) supplemental files (e.g. additional tables, sometimes in PDF) and (c) textual statements about data availability (e.g. ‘available upon request’).
As datasets from category 2b and 2c often required additional effort before they could be used in calculations, we limited our data collection to categories 1 and 2a, which led to the task of identifying URL links from PubMed full-text articles. Of course, external URL links in PMC articles do not always refer to datasets. Therefore, we developed a process that combines automatic extraction with manual review, to identify dataset links mentioned in articles. We first downloaded the full texts of 12 324 PMC articles in XML format using E-Fetch queries [8]. All URLs tagged with the markup ‘ext-link’ were then automatically extracted from articles. This included URLs both in the main text and in the citations. These URLs then underwent a normalization step, where extensions like ‘HTTP’ and ‘htm’ were removed, which resulted in a list of 23 467 URLs in total. We then manually reviewed all of them and identified 144 links directing to actual datasets. We noticed that one single dataset can be associated with multiple links. For example, the Johns Hopkins University Dashboard [9] was cited in articles using four different URLs. After merging these different data links that directed to the same dataset, we obtained 128 unique datasets from the verified data links. The complete process of extracting COVID-19 publications from LitCovid and extracting datasets mentioned in full-text COVID-19 publications in PMC was described in Figure 1.
Figure 1.
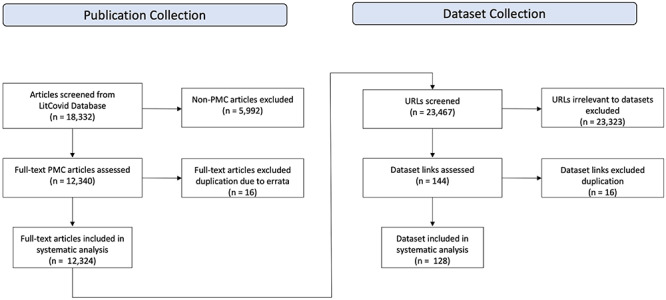
The workflow of screening and collecting publications and datasets from 18 332 PMC articles.
COVID-19 dataset review and analysis
For each of the 128 COVID-19 datasets, we manually reviewed its web pages. We extracted information for 10 descriptive variables: (1) type of the dataset (e.g. epidemiological or genomic data), (2) geographic region where the data were collected, (3) whether the dataset was immediately downloadable, (4) file format (e.g. CSV), (5) where the dataset was hosted, (6) whether the data were updated regularly, (7) the type of license used, (8) whether the metadata were explicitly provided, (9) whether there was a PMC paper describing the creation of the dataset and (10) the number of times the dataset was cited by PMC articles (either via URL links or via articles). The definitions and examples of values for the 10 variables are shown in Table 1.
Table 1.
Descriptions and examples of metadata variables collected for each dataset
| Question | Variable | Description | Examples |
|---|---|---|---|
| Content | Data type | Types of dataset | Epidemiological, clinical, etc. |
| Geographic region | The region from where the data were collected | Worldwide, China, United States, etc. | |
| Accessibility | Download | Can user download the dataset | Immediately downloadable versus Request needed |
| Data format | Format of the dataset | CSV, XLSX, etc. | |
| Data hosting | Data repository where the dataset was hosted | GitHub, Mendeley, etc. | |
| Data update frequency | Whether the dataset was being updated regularly and the last date of update | Regularly updated versus One time only | |
| License | The type of license used | CC BY 4.0, MIT, etc. | |
| Metadata availability | Whether the metadata are provided and the metadata format | Machine readable, unstructured or unavailable | |
| Citation | Dataset article | Whether there was a PMC paper that described the dataset | The JHU dataset was described in the PMID 32087114 article |
| Citation count | The number of times that the dataset was cited by PMC articles (either via URL links or via data articles) | The JHU dataset was cited by 454 PMC articles |
JHU, Johns Hopkins University; CSV, comma separated values; XLSX, Microsoft Excel Open XML Spreadsheet; CC BY 4.0, Creative Commons Attribution 4.0; MIT, The MIT License.
Results
Among 12 324 PMC articles screened, 249 papers included Data Availability Statement sections, and 23 papers provided valid online data sources. Of the papers without the Data Availability Statement (12 075), 3486 papers contained at least one dataset link in the full text. The proportion of COVID-19 articles in PMC that provided at least one dataset link was 28.5% (3509/12 324). In total, 128 unique dataset links were mentioned in 12 324 COVID-19 articles in PMC.
Q1. Content
Data types
Table 2 presents the distribution of types of datasets. As expected, epidemiological datasets (N = 69; 53.9%) constituted the largest portion of our dataset collection. Some of them were created by governmental sources and others by independent statistic suppliers and were aimed at tracking the latest COVID-19 case updates, including confirmed cases, death cases, recovered cases and the number of tests conducted [9–13, 18–23, 31–50, 53–57, 61, 66, 67, 74–76, 132]. Some epidemiological studies focused on the modeling [15, 16, 26, 29, 51, 58, 64, 135] and prediction of transmission patterns [24, 30, 52, 60, 63, 65, 70–73]. Of all datasets, 14.8% provided COVID-19 genome [76–91, 93] or protein sequences [92, 94, 95, 98]. Clinical datasets (N = 15; 11.7%) largely concentrated on three aspects: (1) incubation period as well as other clinical characteristics of COVID-19 patients [96, 104, 107], (2) potential treatments such as vaccines [97, 105] and medications [99] and (3) imaging datasets (N = 3; 2.3%) contained chest computed tomography (CT) images for COVID-19 patients plus others [110–112]. Mobility datasets (N = 7; 5.5%) used track movements trends over time by geography were also present [114–120]. Social studies datasets (N = 4; 3.1%) gathered information about people’s responses when facing the pandemic [121–124]. LitCovid [2] and CORD-19 [113] were the two major COVID-19 literature databases that appeared in PMC articles. Health administration data [100–103, 108] mainly describe hospital capacities, for example the number of ventilators [103] and ICU beds [108] available in hospitals. Other non-biomedical datasets (N = 10; 7.8%), such as climate [133, 134], economic [127], geographical [128] and population data [129, 131], were also discovered in articles investigating the effects of disease transmission and long-term impacts of the pandemic.
Table 2.
The distribution of data types among 128 COVID-19 datasets
| Data type | Number | Percent |
|---|---|---|
| Epidemiology | 69 | 53.9% |
| Genomics | 19 | 14.8% |
| Clinical | 15 | 11.7% |
| Imaging | 3 | 2.3% |
| Mobility | 7 | 5.5% |
| Social science | 4 | 3.1% |
| Healthcare administration | 2 | 1.6% |
| Literature | 2 | 1.6% |
| Other | 10 | 7.8% |
Note: Imaging is considered as a subset of clinical datasets.
Geographic region
Figure 2 illustrates the geographic regions that the datasets covered. More than half of the datasets (N = 68; 53.1%) incorporated data from around the world. From the total, 18 (14.1%) datasets involved data from China. Multiple datasets related to epidemiology were reported in the United States [66, 69, 90, 102], United Kingdom [40, 54, 62] and India [35, 36, 57] as the coronavirus diseases spread to these countries. Aside from country-level data, Africa [59] and Europe [109] also created datasets covering these entire continents. There were also smaller datasets that covered only states [39, 47], counties [56] and cities [14, 16, 17, 43, 53]. Such datasets were often created by local health departments and incorporated detailed COVID-19 patients’ demographic breakdowns.
Figure 2.
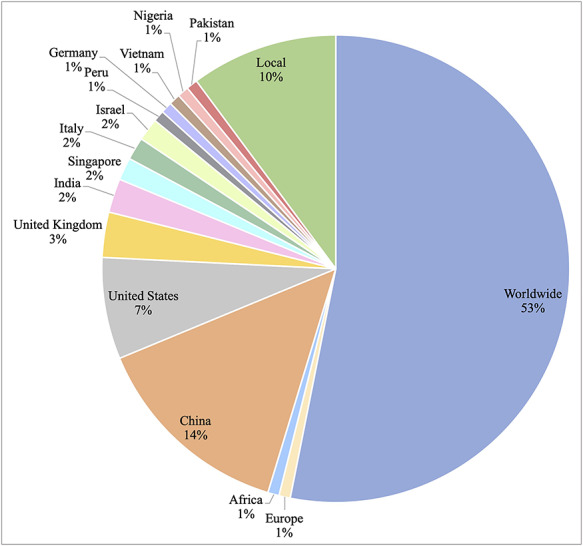
Distribution of geographic regions where COVID-19 datasets were collected.
Q2. Accessibility
Download
Among the 128 datasets in our study, 20 datasets did not provide clear downloading information. Users who wish to use these datasets need to contact the owners for download instructions. Therefore, we marked the accessibility of such datasets as ‘request needed’. The remaining 108 (84.4%) datasets were instantly downloadable. Registrations prior to accessing the data are required for 9 out of 108 downloadable datasets.
Data format
Of 108 datasets that could be downloaded instantly, 19 were available to download in multiple formats. CSV (N = 53; 49.1%), XLSX (N = 27; 25.0%) and JSON (N = 10; 9.3%) were three popular formats in dataset exchange. Almost all genetic studies shared data in FASTA. RDS and RDA were two of the common data formats in studies that utilized the R programming language [28, 60, 68, 106, 108, 126]. Imaging datasets typically shared CT images as JPG files. Datasets of protein structures offered data in PDB files [92, 95]. GeoTiFF files were provided in a worldwide population dataset that allowed the data to be projected onto a geographical map [129].
Figure 3.
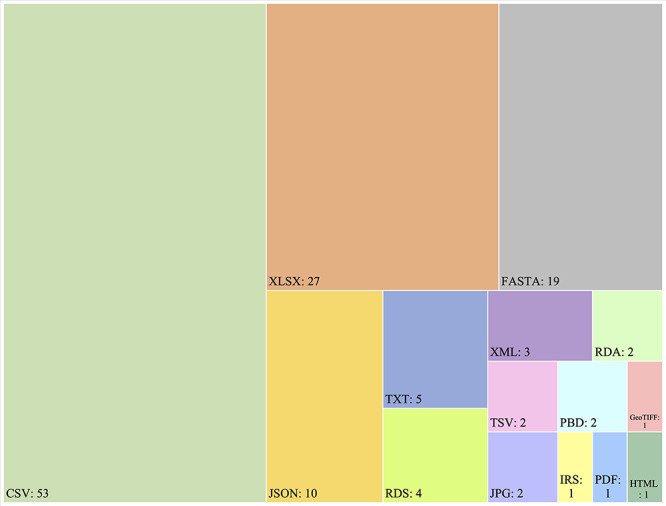
Data formats used by 108 downloadable datasets, among 128 COVID-19 datasets.
Data hosting
As shown in Table 3 below, the most popular data repository is GitHub, incorporating 57 (44.5%) datasets. Of all, six (4.7%) datasets were stored on Mendeley Data, a cloud-based repository for research data from scholarly articles. Individual webpages (N = 55; 43.0%) referred to those datasets accessible only via stand-alone websites, in comparison with those deposited on established data repositories.
Table 3.
List of data repositories used by datasets in this study
| Repository | Number | Percent |
|---|---|---|
| GitHub | 57 | 44.5% |
| Google drive | 7 | 5.5% |
| Mendeley | 6 | 4.7% |
| Kaggle | 3 | 2.3% |
| Individual web page | 55 | 43.0% |
Data update frequency
More than half (N = 74; 57.8%) of the datasets were being updated regularly (often daily or weekly). If data depositors did not offer any information regarding the updating frequency, we treated those datasets as not being updated on a regular basis. We recorded the date of the last update on those datasets. Figure 4 illustrates the number of datasets that stopped updating in each month.
Figure 4.
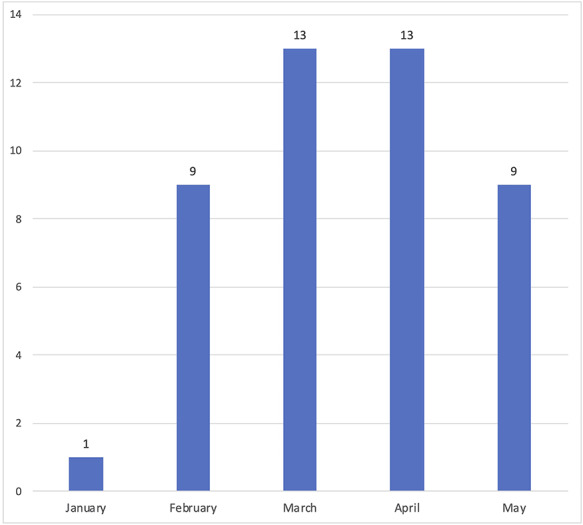
The time of last updates for datasets that were not being updated regularly, among 128 COVID-19 datasets.
License
Table 4 showed the statistics for data licensing. Among the 128 datasets we collected, 39 (30.5%) datasets clearly specified data licenses to allow permitted use of datasets. The COVID-19 Image Data Collection [111] used multiple licenses for different subsets of data. 37.5% (N = 48) stated their own terms and policies for data usage in detail online. 8.6% (N = 11) datasets require users to cite their associated papers when using the data but do not offer other information on data sharing and usage. 23.4% (N = 30) datasets do not release any information regarding data usage.
Table 4.
The number of times that a data license is used in dataset collection
| License | Number | Percent |
|---|---|---|
| MIT | 12 | 9.4% |
| Creative Commons Attribution 4.0 | 12 | 9.4% |
| GNU General Public License v3.0 | 7 | 5.5% |
| Apache license 2.0 | 4 | 3.1% |
| Creative Commons Zero v1.0 Universal | 3 | 2.3% |
| Creative Commons Attribution-NonCommercial-ShareAlike 4.0 | 2 | 1.6% |
| Mozilla Public License 2.0 | 1 | 0.8% |
| Self-defined data usage policy | 48 | 37.5% |
| Citation required | 11 | 8.6% |
| Unavailable | 30 | 23.4% |
Note: One dataset [111] used multiple licenses, thus percentages in this table may not add up. Self-defined data usage: data owners defined their own data usage policy; citation required: data owners only require users to cite their associated papers when using the data.
Metadata availability
Of 108 datasets that are immediately downloadable, 77.8% (N = 84) provide metadata in machine readable formats. Several datasets [40, 74, 78, 125, 130] and data deposited on established data repositories (GitHub, Mendeley and Kaggle) offer application programming interfaces (API) to automatically retrieve metadata. 9.3% (N = 10) datasets provide metadata in free text, which includes information like dataset names, data owners and data description. 13.0% (N = 14) datasets do not release any information on metadata.
Q3. Citation
Dataset article availability
41.4% (N = 53) datasets were described with details in publications on PMC. Of the 53 articles describing datasets, 5 articles described extensively the purpose and techniques of building COVID-19 databases. The main purpose of the remaining 48 articles was to carry out modeling, prediction or other types of analysis in diverse domains, with some description about datasets in the study. These were often the datasets that were not updated on a regular basis: those data were collected, standardized and maintained by the authors themselves for the specific studies.
Citation count
Figure 5 demonstrates the number of citations for each dataset. Typically, a dataset can be cited in two ways in articles: (1) as a URL in the full text and (2) as an article that describes the dataset. It is possible for an article to cite both the URL and the article of the dataset. The number shown in Figure 5 is for the overall citations (both articles and URLs), in which the duplicated citations were removed. The number of citations across different datasets varied heavily. The dataset available in the John Hopkins University Dashboard [9] was the most popular dataset and was cited 454 times. Of the top 10 datasets, 9 were from the epidemiology domain. However, a low number of citations do not necessarily indicate that the dataset has little impact and may just reflect the fact that they did not have enough time to accrue citations yet (i.e. more recently published datasets).
Figure 5.
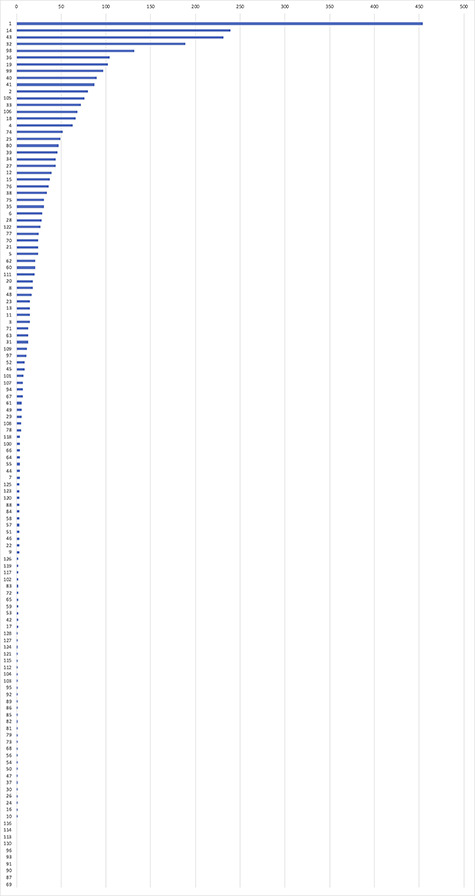
Number of citations for each dataset. The horizontal axis indicates the number of citations of a dataset. The vertical axis label corresponds to the dataset ID in our dataset summary list (included in the Supplementary Data available online at https://academic.oup.com/bib).
Table 5 presents the top 10 cited datasets in our study. The John Hopkins University Dashboard [9] had a large number of citations both as an online data link and a publication. Worldometers [33] and CDC [31] are high-impact data sources for COVID-19 case update and cited frequently as external data links. They are used in the Johns Hopkins University Dashboard but, since they do not accrue citations indirectly, their impact may be underestimated. The remaining seven datasets were almost all cited as articles published on PubMed and had none or few URL citations.
Table 5.
Top 10 cited datasets
| Dataset | Overall citations | URL citations | Article citations |
|---|---|---|---|
| John Hopkins University Dashboard [6] | 454 | 416 | 275 |
| Real-time estimation of the novel coronavirus incubation time [93] | 239 | 0 | 239 |
| Worldometers [30] | 231 | 231 | 0 |
| Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) [22] | 189 | 0 | 189 |
| Estimates of the severity of coronavirus disease 2019: a model-based analysis [60] | 132 | 0 | 132 |
| Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts [25] | 104 | 0 | 104 |
| Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus [14] | 102 | 1 | 102 |
| Early dynamics of transmission and control of COVID-19: a mathematical modelling study [61] | 97 | 0 | 97 |
| The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak [27] | 90 | 0 | 90 |
| CDC [28] | 87 | 87 | 0 |
Note: The number may not add up to the number of overall citations as we merged URL citations and article citations from the same article.
Discussion
Although no extensive analyses have been carried out on availability, accessibility and type of COVID-19 datasets, discussion on the collection and sharing of COVID-19 data has received great attention among the scientific community: Alamo et al. [136] highlighted a variety of significant open data sources and evaluated the limitations and readability of available data. They concluded that notable progress was achieved by certain scientific communities, particularly among epidemiologists, healthcare specialists, the machine learning community and data scientists. Several studies also reviewed and explored available COVID-19 data in specific domains. Kalkreuth and Kaufmann [137] reviewed publicly available medical imaging resources for COVID-19 cases worldwide. Rubin [138] reported on recent progress in collecting data of ventilated patients confirmed with COVID-19. Robinson and Yazdany [139] described an initiative to collect data about COVID-19 patients with rheumatic diseases. Khalatbari-Soltani et al. [140] listed a series of important socioeconomic characteristics often overlooked when collecting and reporting social science data related to the pandemic.
In our study, we took a different approach and conducted a systematic review on COVID-19 literature in PMC to identify associated datasets. A number of interesting findings were identified through our analysis. First, although PMC implemented the Data Availability Statement section, the percentage of articles that explicitly provided such information was about 2% (249/12 324) only, indicating a low adoption rate. Nevertheless, there were 3509 (about 28.5%) papers that provided at least one URL link to datasets used in the study, demonstrating a significant portion of researchers are aware of the importance of sharing data. Additionally, all 128 datasets identified in PMC articles allowed users to access their data, and 84.4% (108/128) were available for immediate download, indicating the level of openness of data sharing in COVID-19 research. Epidemiology datasets constituted more than half of our dataset collection, while imaging datasets accounted for 2.3%, indicating the need to develop more datasets for the latter and for related domains, which will probably require worldwide collaboration in order to grow to the same size as epidemiology datasets. As for data format, although FAIR [5] recommends the RDF (Resource Description Framework) format, no dataset in this study has adopted RDF, probably because common machine-readable formats such as CSV, JSON and TXT are easier to understand. We observed two major types of practices in licenses of data usage. Data owners who use established data repositories often use a variety of existing data licenses to grant data usage and sharing. On the other hand, data owners who publish datasets on individual webpages prefer to specify their own terms and policies. Overall, 76.6% (98/128) data owners allow non-commercial use of data and specify the degree of openness by releasing data usage policies.
The data update frequency relied heavily on the objectives of creating the dataset. Among 75 datasets only available as online sources, the majority of them were updated regularly for public uses. However, for 41.4% (53/128) datasets that are associated with publications, the authors collected and maintained datasets themselves for different purposes. Five articles aimed at describing how the COVID-19 databases were built, and they discuss data collection, storage and visualization. The remaining 48 articles focused on modeling, predictions or other analysis related to COVID-19. The authors of these analysis articles kept not only data but also codes and tools they used in their own studies. The datasets mentioned in these articles represent the collection of raw data that authors used as input for their analysis. Such data are often limited within a period of time and contain a relatively small number of cases.
We observed two approaches for citing datasets: (1) URL citations: citing URLs that led to the data sources and (2) Article citations: citing the article that describes the dataset. After examining the articles that cited datasets in the full text, we also discovered two major purposes of citing datasets: (1) citing a dataset as the data source used in the study and (2) citing a dataset as a general reference. Researchers are typically more likely to have used the dataset if they cite it directly as a URL. On the other hand, when citing a dataset as an article, the authors are more likely to mention it as a general reference instead of citing the data sources. This suggests that a larger number of URL citations to a dataset indicate its higher reuse. However, we also saw that datasets that aggregate data from several sources can be popular and be highly cited, but the data sources they use may not always receive citations. This indicates that we may consider indirect citations when assessing the true impact of a dataset in terms of its utility. Additionally, if a dataset is associated with a dedicated description paper, e.g. the John Hopkins University Dashboard [9] or the Epidemiological Data from the nCoV-2019 Outbreak [18], other papers that used the dataset may cite it as both URLs and papers.
One limitation of this study is that we limited our analysis to full-text articles in PMC. Although PMC is the largest full-text article repository in the biomedical domain, there are still about one-third (5992/18 332) of LitCovid papers that are not included in this study due to unavailability at PMC. Considering that LitCovid collects articles from PubMed only, the actual number of COVID-19 articles that are not included in this study could be even higher. In the future, we plan to look into other sources of full-text articles to study COVID-19 dataset status. Additionally, our study did not take into account high-impact datasets cited often by preprints, such as the Public Coronavirus Twitter Dataset [141]. Furthermore, we reviewed only the URLs extracted from articles, instead of other potential types of references that could be revealed had we reviewed the whole text. There is a chance that we missed data source information stated in plain text. We hope to resolve this problem and to expand the dataset collection by introducing natural language processing techniques in our future studies.
Conclusion
We screened 12 324 COVID-19 related full-text articles in PMC and collected 128 unique dataset URLs. By systematically analyzing the collected datasets in terms of content, accessibility and citation, we observed significant heterogeneity in the way these datasets are mentioned, shared, updated and cited. Those findings on current practice on generating, sharing and citing datasets for COVID-19 research can provide valuable insights for future improvements.
Key Points
128 COVID-19 datasets from 12 324 COVID-19 articles were collected for this systematic review.
We conducted a quantitative analysis of dataset contents, accessibility and citations.
84.4% COVID-19 scholarly datasets are available for immediate download.
The number of dataset URL citations is a valuable indicator of dataset utility.
Supplementary Material
Xu Zuo is a PhD student at School of Biomedical Informatics, The University of Texas Health Science Center at Houston. Her research focuses on clinical natural language processing.
Yong Chen is an associate professor of biostatistics at the University of Pennsylvania. His research interests include evidence synthesis, data integration and real-world evidence.
Lucila Ohno-Machado is a professor of medicine (Biomedical Informatics) at the University of California San Diego. Her research interests include privacy technology, predictive analytics and data sharing.
Hua Xu is a professor at School of Biomedical Informatics, The University of Texas Health Science Center at Houston. His research interests include biomedical text mining and data mining.
Contributor Information
Xu Zuo, School of Biomedical Informatics, The University of Texas Health Science Center at Houston.
Yong Chen, University of Pennsylvania.
Lucila Ohno-Machado, University of California San Diego.
Hua Xu, School of Biomedical Informatics, The University of Texas Health Science Center at Houston.
Data Availability
The original data presented in the study are included in the article/supplementary materials.
Funding
UTHealth CCTS Pilot Project (0015300); National Science Foundation (OIA-1937136).
References
- 1. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. Geneva, Switzerland: World Health Organization; https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (11 March 2020, date last accessed). [Google Scholar]
- 2. Chen Q, Allot A, Lu Z. Keep up with the latest coronavirus research. Nature 2020;579(7798):193. [DOI] [PubMed] [Google Scholar]
- 3. Ohno-Machado L, Xu H. Coronavirus: indexed data speed up solutions. Nature 2020;584:192. doi: 10.1038/d41586-020-02331-3. [DOI] [PubMed] [Google Scholar]
- 4. Forster P, Forster L, Renfrew C, et al. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA 2020;117(17):9241–3. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin D, Crabtree J, Dillo I, et al. The TRUST principles for digital repositories. Sci Data 2020;7:144. doi: 10.1038/s41597-020-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virus Outbreak Data Network (VODAN). https://www.go-fair.org/implementation-networks/overview/vodan/ (27 July 2020, date last accessed).
- 8.Do text mining/retrieving full text. https://www.ncbi.nlm.nih.gov/pmc/tools/get-full-text/ (27 July 2020, date last accessed).
- 9. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020;20(5):533–4. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. European Centre for Disease Prevention and Control : COVID-19 case update worldwide. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases (27 June 2020, date last accessed).
- 11.China Centre for Disease Prevention and Control COVID-19 Dashboard. http://2019ncov.chinacdc.cn/2019-nCoV/ (27 June 2020, date last accessed).
- 12.Online repository (MOBS). https://docs.google.com/spreadsheets/d/e/2PACX-1vQU0SIALScXx8VXDX7yKNKWWPKE1YjFlWc6VTEVSN45CklWWf-uWmprQIyLtoPDA18tX9cFDr-aQ9S6/pubhtml (27 June 2020, date last accessed).
- 13.WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ (27 June 2020, date last accessed).
- 14. Du Z, Wang L, Cauchemez S, et al. Risk for transportation of coronavirus disease from Wuhan to other cities in China. Emerg Infect Dis 2020;26(5):1049–52. doi: 10.3201/eid2605.200146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estimating case fatality ratio of COVID-19 from observed cases outside China. https://github.com/calthaus/ncov-cfr (27 June 2020, date last accessed).
- 16.Analysis of early transmission dynamics of nCoV in Wuhan. https://github.com/zsvizi/corona-virus-2020 (27 June 2020, date last accessed).
- 17. Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill 2020;25(4):2000058. doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu B, MUG K, Open COVID-19 Data Curation Group . Open access epidemiological data from the COVID-19 outbreak. Lancet Infect Dis 2020;20(5):534. doi: 10.1016/S1473-3099(20)30119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singapore Ministry of Health. https://www.moh.gov.sg/covid-19 (27 June 2020, date last accessed).
- 20.COVID19 Outbreak tracking and forecast. https://docs.google.com/spreadsheets/d/1f3LGuqwzegr7ZdGlzPOCDAyFk8RTaLTmMLF_K_5EVCc/edit#gid=783518927 (27 June 2020, date last accessed).
- 21.Our World in Data. https://ourworldindata.org/coronavirus (27 June 2020, date last accessed).
- 22. Italian Civil Protection Department, Morettini M, Sbrollini A, et al. COVID-19 in Italy: dataset of the Italian civil protection department. Data Brief 2020;30:105526.Published 2020 Apr 10. doi: 10.1016/j.dib.2020.105526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latest Situation of Coronavirus Disease (COVID-19) in Hong Kong. https://chp-dashboard.geodata.gov.hk/covid-19/en.html (27 June 2020, date last accessed).
- 24. Wells CR, Sah P, Moghadas SM, et al. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc Natl Acad Sci U S A 2020;117(13):7504–9. doi: 10.1073/pnas.2002616117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020;368(6490):489–93. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kraemer MUG, Yang CH, Gutierrez B, et al. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science 2020;368(6490):493–7. doi: 10.1126/science.abb4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du Z, Xu X, Wu Y, et al. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis 2020;26(6):1341–3. doi: 10.3201/eid2606.200357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 2020;8(4):e488–96. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis 2020;93:284–6. doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chinazzi M, Davis JT, Ajelli M, et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science 2020;368(6489):395–400. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention: Cases, Data, and Surveillance. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/index.html (27 June 2020, date last accessed).
- 32.Bing COVID-19 Tracker. https://github.com/microsoft/Bing-COVID-19-Data (27 June 2020, date last accessed).
- 33.Worldometers. https://www.worldometers.info/coronavirus/ (27 June 2020, date last accessed).
- 34.Pakistan COVID case update. http://covid.gov.pk/stats/pakistan (27 June 2020, date last accessed).
- 35.India COVID-19 Statewise Status. https://www.mygov.in/covid-19/ (27 June 2020, date last accessed).
- 36.Tracking the impact of COVID-19 in India. https://github.com/covid19india/covid19india-react (27 June 2020, date last accessed).
- 37.Oxford COVID-19 Evidence Service. http://www.cebm.net/oxford-covid-19-evidence-service/ (27 June 2020, date last accessed).
- 38.Global Health 5050. https://globalhealth5050.org/covid19/ (27 June 2020, date last accessed).
- 39.Rhode Island COVID-19 response data. https://ri-department-of-health-covid-19-data-rihealth.hub.arcgis.com/ (27 June 2020, date last accessed).
- 40.Oxford COVID-19 Government Response Tracker. https://covidtracker.bsg.ox.ac.uk/ (27 June 2020, date last accessed).
- 41.The COVID Tracking Project. https://covidtracking.com/data (27 June 2020, date last accessed).
- 42.King County COVID-19 data dashboards. https://kingcounty.gov/depts/health/covid-19/data.aspx (27 June 2020, date last accessed).
- 43.Shenzhen COVID case update. https://opendata.sz.gov.cn/data/dataSet/toDataDetails/29200_01503668 (27 June 2020, date last accessed).
- 44.Google News COVID-19. https://news.google.com/covid19/map?hl=en-US&gl=US&ceid=US:en (27 June 2020, date last accessed).
- 45.Peru COVID update. http://covid19.minsa.gob.pe/sala_situacional.asp (27 June 2020, date last accessed).
- 46. Sun K, Chen J, Viboud C. Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: a population-level observational study. Lancet Digit Health 2020;2(4):e201–8. doi: 10.1016/S2589-7500(20)30026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.NYSDOH COVID-19 Tracker. https://covid19tracker.health.ny.gov/views/NYS-COVID19-Tracker/NYSDOHCOVID-19Tracker-Map?%3Aembed=yes&%3Atoolbar=no&%3Atabs=n (27 June 2020, date last accessed).
- 48.SECURE IBD. https://covidibd.org/current-data/ (27 June 2020, date last accessed).
- 49.COVID-19 Projections. http://covid19.healthdata.org (27 June 2020, date last accessed).
- 50.nCoV2019 Live. https://ncov2019.live/ (27 June 2020, date last accessed).
- 51.State-level social distancing policies in response to the 2019 novel coronavirus in the US. https://github.com/COVID19StatePolicy/SocialDistancing (27 June 2020, date last accessed).
- 52. Tian H, Liu Y, Li Y, et al. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science 2020;368(6491):638–42. doi: 10.1126/science.abb6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.NYC Coronavirus Disease 2019 (COVID-19) Data. https://github.com/nychealth/coronavirus-data (27 June 2020, date last accessed).
- 54.Coronavirus (COVID-19) in the UK. https://coronavirus.data.gov.uk/ (27 June 2020, date last accessed).
- 55.Israeli COVID-19 Database. https://data.gov.il/dataset/covid-19 (27 June 2020, date last accessed).
- 56.Amtliches COVID-19 Dashboard. https://info.gesundheitsministerium.at/dashboard_Hosp.html?l=en (27 June 2020, date last accessed).
- 57.COVID-19 cases in the Indian health system. Available at: https://docs.google.com/spreadsheets/d/11ng2dly8jjc6RZ-aiT7xfQNKiD6MDPUwMXOnOJ0H7ME/edit#gid=0 (27 June 2020, date last accessed).
- 58. Millett GA, Jones AT, Benkeser D, et al. Assessing differential impacts of COVID-19 on black communities. Ann Epidemiol 2020;47:37–44. doi: 10.1016/j.annepidem.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pearson CA, Van Schalkwyk C, Foss AM, et al. Projected early spread of COVID-19 in Africa through 1 June 2020. Euro Surveill 2020;25(18):2000543. doi: 10.2807/1560-7917.ES.2020.25.18.2000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jarvis CI, Van Zandvoort K, Gimma A, et al. Quantifying the impact of physical distance measures on the transmission of COVID-19 in the UK BMC Med. 2020;18(1):124. doi: 10.1186/s12916-020-01597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Data on the 2019 Novel Coronavirus Outbreak. https://github.com/eebrown/data2019nCoV/ (27 June 2020, date last accessed).
- 62. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020;369(6501):297–301. doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis 2020;20(6):669–77. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kucharski AJ, Russell TW, Diamond C, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis 2020;20(5):553–8. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chakraborty T, Ghosh I. Real-time forecasts and risk assessment of novel coronavirus (COVID-19) cases: a data-driven analysis. Chaos Solitons Fractals 2020;135:109850. doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.NY Times Coronavirus (Covid-19) Data in the United States. https://github.com/nytimes/covid-19-data (27 June 2020, date last accessed).
- 67. Rossman H, Keshet A, Shilo S, et al. A framework for identifying regional outbreak and spread of COVID-19 from one-minute population-wide surveys. Nat Med 2020;26(5):634–8. doi: 10.1038/s41591-020-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abbott S, Hellewell J, Munday J, et al. The transmissibility of novel coronavirus in the early stages of the 2019-20 outbreak in Wuhan: exploring initial point-source exposure sizes and durations using scenario analysis. Wellcome Open Res 2020;5:17. doi: 10.12688/wellcomeopenres.15718.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baker MG, Peckham TK, Seixas NS. Estimating the burden of United States workers exposed to infection or disease: a key factor in containing risk of COVID-19 infection. PLoS One 2020;15(4):e0232452. doi: 10.1371/journal.pone.0232452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ganyani T, Kremer C, Chen D, et al. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill 2020;25(17):2000257. doi: 10.2807/1560-7917.ES.2020.25.17.2000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Russell TW, Hellewell J, Jarvis CI, et al. Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill 2020;25(12):2000256. doi: 10.2807/1560-7917.ES.2020.25.12.2000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abdulmajeed K, Adeleke M, Popoola L. Online forecasting of COVID-19 cases in Nigeria using limited data. Data Brief 2020;30:105683. doi: 10.1016/j.dib.2020.105683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reis RF, Melo QB, Oliveira CJ, et al. Characterization of the COVID-19 pandemic and the impact of uncertainties, mitigation strategies, and underreporting of cases in South Korea, Italy, and Brazil. Chaos Solitons Fractals 2020;136:109888. doi: 10.1016/j.chaos.2020.109888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.1Point3Acres. https://coronavirus.1point3acres.com/ (27 June 2020, date last accessed).
- 75.Day level information on covid-19 affected cases. https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset (27 June 2020, date last accessed).
- 76.Kaggle-Coronavirus Disease 2019 cases in Italy. https://www.kaggle.com/sudalairajkumar/covid19-in-italy (27 June 2020, date last accessed).
- 77.GISAID. https://www.gisaid.org/ (27 June 2020, date last accessed).
- 78.Genbank. https://www.ncbi.nlm.nih.gov/genbank/ (27 June 2020, date last accessed).
- 79.NextStrain. https://github.com/nextstrain/ncov (27 June 2020, date last accessed).
- 80.China National Genomics Data Center. https://bigd.big.ac.cn/databases?lang=en (27 June 2020, date last accessed).
- 81. Qiang XL, Xu P, Fang G, et al. Using the spike protein feature to predict infection risk and monitor the evolutionary dynamic of coronavirus. Infect Dis Poverty 2020;9(1):33. doi: 10.1186/s40249-020-00649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Barbosa RM, Fernandes MAC. Chaos game representation dataset of SARS-CoV-2 genome. Data Brief 2020;30:105618. doi: 10.1016/j.dib.2020.105618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Alakwaa FM. Repurposing didanosine as a potential treatment for COVID-19 using single-cell RNA sequencing data. mSystems 2020;5(2):e00297–20. doi: 10.1128/mSystems.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim D, Lee JY, Yang JS, et al. Cell 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 2020;9(1):761–70. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lukassen S, Chua RL, Trefzer T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39(10):e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181(5):1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.COVID19 Virtual BioHackathon 2020. https://github.com/virtual-biohackathons/covid-19-bh20/wiki (27 June 2020, date last accessed).
- 89. Lu J, Plessis L, Liu Z, et al. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 2020;181(5):997–1003.e9. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fauver JR, Petrone ME, Hodcroft EB, et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 2020;(5):181, 990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.ARTIC nanopore protocol for nCoV2019 novel coronavirus. https://github.com/artic-network/artic-ncov2019 (27 June 2020, date last accessed).
- 92.RCSB Protein Data Bank. https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true (27 June 2020, date last accessed).
- 93.NCBI Virus. https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=Severe%20acute%20respiratory%20syndrome%20coronavirus%202%20taxid:2697049 (27 June 2020, date last accessed).
- 94.Broad Terra. https://app.terra.bio/ (27 June 2020, date last accessed).
- 95.Collection of 3D Print Models of SARS-CoV-2 virions and proteins. https://3dprint.nih.gov/niaid/sars-cov-2 (27 June 2020, date last accessed).
- 96. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020;172(9):577–82. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ton AT, Gentile F, Hsing M, et al. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inform 2020;39(8):e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Global COVID19 Telehealth Directory. https://docs.google.com/spreadsheets/d/1XMsJJIduO6yI_GEo1Vy_b_SXoz9YwbgtEL63-siNS_Q/edit#gid=0 (27 June 2020, date last accessed).
- 101.COVID-19 Cases on ECMO in the ELSO Registry. https://www.elso.org/Default.aspx?TabID=576 (27 June 2020, date last accessed).
- 102. Moghadas SM, Shoukat A, Fitzpatrick MC, et al. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc Natl Acad Sci U S A 2020;117(16):9122–6. doi: 10.1073/pnas.2004064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.COVID-19 Ventilator Projects and Resources with FAQs. https://github.com/PubInv/covid19-vent-list (27 June 2020, date last accessed).
- 104. Wang S, Zha Y, Li W, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J 2020;56(2):2000775. doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lee CH, Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Res 2020;9:145. doi: 10.12688/f1000research.22507.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020;181(4):905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Linton NM, Kobayashi T, Yang Y, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med 2020;9(2):538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Alban A, Chick SE, Dongelmans DA, et al. ICU capacity management during the COVID-19 pandemic using a process simulation. Intensive Care Med 2020;46(8):1624–6. doi: 10.1007/s00134-020-06066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.The European Data Portal. https://data.europa.eu/euodp/es/data/ (27 June 2020, date last accessed).
- 110. Li L, Qin L, Xu Z, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 2020;296(2):E65–71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.COVID-19 image data collection. https://github.com/ieee8023/covid-chestxray-dataset (27 June 2020, date last accessed).
- 112.Chest Xray Pneumonia. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia (27 June 2020, date last accessed).
- 113. Lu Wang L, Lo K, Chandrasekhar Y, et al. CORD-19: the Covid-19 open research dataset. ArXiv Preprint 2020. arXiv: 2004.10706v2. Published 22 April 2020. [Google Scholar]
- 114.Tencent Heat. https://heat.qq.com/ (27 June 2020, date last accessed).
- 115.Baidu Qianxi. http://qianxi.baidu.com/ (27 June 2020, date last accessed).
- 116.VariFlight. https://data.variflight.com/ (27 June 2020, date last accessed).
- 117.Flightradar24. https://www.flightradar24.com/ (27 June 2020, date last accessed).
- 118.Google COVID-19 Community Mobility Reports. https://www.google.com/covid19/mobility/ (27 June 2020, date last accessed).
- 119.Apple Mobility Trends Reports. https://www.apple.com/covid19/mobility (27 June 2020, date last accessed).
- 120. Ribeiro-Dantas MDC, Alves G, Gomes RB, et al. Dataset for country profile and mobility analysis in the assessment of COVID-19 pandemic. Data Brief 2020;31:105698. doi: 10.1016/j.dib.2020.105698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bento AI, Nguyen T, Wing C, et al. Evidence from internet search data shows information-seeking responses to news of local COVID-19 cases. Proc Natl Acad Sci U S A 2020;117(21):11220–2. doi: 10.1073/pnas.2005335117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Aguilar-Gallegos N, Romero-García LE, Martínez-González EG, et al. Dataset on dynamics of coronavirus on twitter. Data Brief 2020;30:105684. doi: 10.1016/j.dib.2020.105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Huynh TLD Data for understanding the risk perception of COVID-19 from Vietnamese sample. Data Brief 2020;30:105530. doi: 10.1016/j.dib.2020.105530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Open ICPSR. https://www.openicpsr.org/openicpsr/covid19 (27 June 2020, date last accessed).
- 125.CEIC Data. https://www.ceicdata.com/en (27 June 2020, date last accessed).
- 126.GADM Data. https://gadm.org/index.html (27 June 2020, date last accessed).
- 127.The World Bank. http://datatopics.worldbank.org/universal-health-coverage/coronavirus/ (27 June 2020, date last accessed).
- 128.National Earth System Science Data Center. http://www.geodata.cn/ (27 June 2020, date last accessed).
- 129.Gridded Population of the World. https://sedac.ciesin.columbia.edu/data/set/gpw-v4-population-density-rev11/data-download (27 June 2020, date last accessed).
- 130.OECD Data. https://data.oecd.org/ (27 June 2020, date last accessed).
- 131.United Nations Population Fund. https://www.unfpa.org/data/world-population-dashboard (27 June 2020, date last accessed).
- 132.City Health Dashboard. https://www.cityhealthdashboard.com/ (27 June 2020, date last accessed).
- 133.National Centers for Environmental Information. https://www.ncei.noaa.gov (27 June 2020, date last accessed).
- 134.ECMWF. https://cds.climate.copernicus.eu/ (27 June 2020, date last accessed).
- 135. Gostic K, Gomez AC, Mummah RO, et al. Estimated effectiveness of symptom and risk screening to prevent the spread of COVID-19. Elife 2020;9:e55570. doi: 10.7554/eLife.55570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Alamo T, Reina DG, Mammarella M, et al. Open data resources for fighting COVID-19. arXiv Preprint 2020. arXiv: 2004.06111. . [Google Scholar]
- 137. Kalkreuth R, Kaufmann P. COVID-19: a survey on public medical imaging data resources. arXiv Preprint 2020. arXiv: 2004.04569. [Google Scholar]
- 138. Rubin R. Global Effort to Collect Data on Ventilated Patients With COVID-19. JAMA 2020;323:2233–4. doi: 10.1001/jama.2020.8341. [DOI] [PubMed] [Google Scholar]
- 139. Robinson PC, Yazdany J. The COVID-19 global rheumatology alliance: collecting data in a pandemic. Nat Rev Rheumatol 2020;16(6):293–4. doi: 10.1038/s41584-020-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Khalatbari-Soltani S, Cumming RC, Delpierre C, et al. Importance of collecting data on socioeconomic determinants from the early stage of the COVID-19 outbreak onwards. J Epidemiol Community Health 2020;74(8):620–3. doi: 10.1136/jech-2020-214297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chen E, Lerman K, Ferrara E. Tracking social media discourse about the COVID-19 pandemic: development of a public coronavirus twitter data set. arXiv Preprint 2020. arXiv: 2003.07372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data presented in the study are included in the article/supplementary materials.


