Abstract
We present an approach for preparing cryoEM grids to study short-lived molecular states. Using piezo electric dispensing, two independent streams of ~50 pL sample drops are deposited within 10 ms of each other onto a nanowire EM grid surface, and the mixing reaction stops when the grid is vitrified in liquid ethane, ~100 ms later. We demonstrate this approach for four biological systems where short-lived states are of high interest.
Keywords: time-resolved, short-lived molecular states, cryoEM vitrification, piezo dispensing, nanowire grids
Cryo electron microscopy (cryoEM) has the distinct advantage of being able to capture a wide variety of conformational states of macromolecules in solution. Changes in conformational states can be triggered by a variety of biological reactions; adding a ligand to an enzyme, mixing together components of a multimolecular machine, or adding energy in the form of ATP or GTP. Conformational changes are often transient but can be trapped by vitrification at specific time points following the initiation of the reaction and imaged using electron microscopy, a process which has been loosely referred to as “time-resolved cryoEM”1 when applied to the study of a conformational process occurring on the millisecond time scale. For much slower processes, standard vitrification or negative staining at fixed time points (on the order of seconds to minutes) serves the same goal2.
Possibly the earliest approach to time-resolved cryoEM was by Berriman and Unwin3 where one reactant was sprayed from an atomizer placed just above the cryogen onto a second reactant already present as a thin aqueous film on an EM grid plunging towards a cryogen at high speed. The potential of this method was demonstrated by spraying acetylcholine onto acetylcholine receptor tubes4. Later, the White group described a computer-controlled, “spraying-mixing” device and observed the interaction of myosin sprayed onto F-actin filaments5. Although the technique can achieve time resolutions as low as 2 ms6, mixing is not uniform across the grid and requires fiducial markers in the sprayed solution to identify regions where mixing occurred. A third approach involved exposing crystals of bacteriorhodopsin on an EM grid to variable periods of light illumination, thus inducing a conformational change just prior to vitrification7. These approaches generated significant interest, but widespread adoption was restricted by practical limitations. An alternative “mixing-spraying” approach8 is based on the spraying-mixing device described above, but mixes the two samples prior to spraying onto a dry grid plunging towards the cryogen. A microfabricated device enables very fast mixing through chaotic advection9 within a fixed reaction chamber length to control the reaction time prior to pneumatic spraying. The system, recently updated to allow either mixing-spraying or spraying-mixing, is capable of reaction times of as low as ~10 ms and has been used in a variety of time-resolved experiments to study the mechanics of ribosomes10. Applying this approach to proteins other than the ribosome may present difficulties, because of the requirement for fairly large (~30 μL) volumes of protein sample. In addition, the thickness of the vitreous ice layer varies with droplet size and the spreading of the droplet on the grid, reducing the efficiency of data collection.
We have developed a new approach to the spraying-mixing method based on the Spotiton robot11,12 that uses a piezo dispensing tip to apply a stream of ~50 pL droplets onto a nanowire (“self-wicking”) grid13 as it rapidly speeds past on its way to vitrification, resulting in a stripe of ice of fairly uniform thickness across each grid. The fast spot-to-plunge time can reduce the deleterious effects of the air-water interface and has been used to prepare grids for a wide variety of protein samples14–17. By adding a second piezo dispensing tip to the device, we can deliver a second stream of droplets onto the first stream within 10 ms of it being deposited. The two sample volumes mix on the grid as the bulk volume is wicked away and spread out to a thin film by the capillary action of the nanowires. Currently, the dual dispenser system can produce useful grids across a range of mixing time intervals from ~90–500 ms. Changes to the physical design of the machine and improving the wicking capacity of the nanowire grids should allow for reducing the lower limit to ~50 ms.
Here we describe the method and demonstrate its efficacy and value for four biological systems where short-lived states are of high interest: (i) binding of ribosomal subunits; (ii) binding of promoter DNA to RNA polymerase; (iii) binding of Ca2+ to a potassium channel followed by a conformational change; (iv) conformational rearrangements of dynamin lipid tubes driven by GTP hydrolysis. The biological results reveal interesting changes in conformations, but we note that further work will be required to determine if the kinetics and thermodynamics are altered by adsorption to the air-water interface, an inevitable result of vitrification of thin films18.
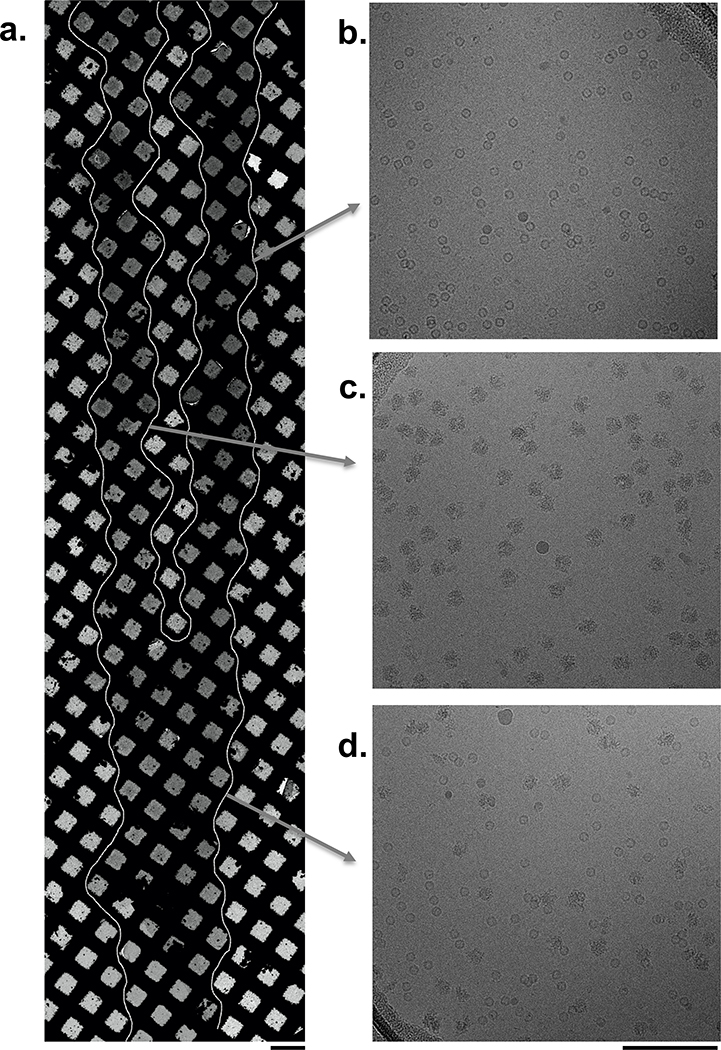
Volumes on the order of 50 pL have been shown to mix completely within ~10 ms when brought together in mid-air just before colliding with a surface19 and thus good mixing of the drops on the nanowire surface is expected. Nevertheless, as a first proof of principle, we mixed two test samples, apoferritin and 70S ribosomes. In practice, one can control the extent of sample stream overlap either by adjusting the dispenser head alignment or the grid acceleration rate (see Methods). Figure 1 shows an example where two samples (Table 1) were deposited onto the grid by setting the second stream of sample droplets to initially overlap the first stream at the leading edge of the grid and then separate from it by 1–3 squares near the grid’s trailing edge towards the end of sample deposition. This method allows comparison of the unmixed control and mixed experiments on the same grid. As observed in the images in Figure 1, in the non-overlapping regions we see apoferritin or 70S ribosomes in high concentrations and well distributed in the vitreous ice of the individual separated stripes, whereas in the overlapping area, we observe both particles well mixed. Except for this proof of principle, for all other experiments the dispenser heads were aligned so that the samples overlap entirely across the grid.
Figure 1: Apoferritin and 70S ribosomes were used as a proof of principle to illustrate mixing on the nanowire grids.
(a) Overview of the vitrified grid showing the sample streams merged at the leading edge (bottom) and separated at the trailing edge (top). Squares containing vitrified ice are indicated by a white outline. Scale bar, 100 μm. Micrographs obtained from the indicated regions show either only (b) apoferritin or (c) 70S ribosomes or (d) a mix of both samples and are representative of approximately 470 images each. Scale bar, 100 nm.
Table 1:
Spotiton sample preparation conditions
| Experiment | Sample (concentration in dispenser) | |
|---|---|---|
| ApoF + 70S mixed | Tip 1 | Apoferritin (2.3 mg/ml) |
| Tip 2 | 70S ribosomes (1mg/ml) | |
| 50S + 30S mixed | Tip 1 | 50S (1.4 μM) |
| Tip 2 | 30S (1.4 μM) | |
| 50S or 30S only | Tip 1 | 50S or 30S (1.4 μM) |
| Tip 2 | Buffer | |
| MthK + Ca2+ mixed | Tip 1 | MthK (12 mg/ml), Fos8-F (3 mM) |
| Tip 2 | Ca2+ (30 mM), Fos8-F (3 mM) | |
| MthK only | Tip 1 | MthK (12 mg/ml), Fos8-F (3 mM) |
| Tip 2 | Buffer, Fos8-F (3 mM) | |
| RNAP + DNA mixed | Tip 1 | DNA (24 uM), beta-OG (0.35%) |
| Tip 2 | RNAP (8 mg/ml), beta-OG (0.35%) | |
| RNAP only | Tip 1 | Buffer, beta-OG (0.35%) |
| Tip 2 | RNAP (8 mg/ml), beta-OG (0.35%) | |
| Dynamin tubes + GTP mixed | Tip 1 | Dynamin tubes |
| Tip 2 | GTP (2 mM & 4 mM) | |
| Dynamin only | Tip 1 | Dynamin tubes |
| Tip 2 | Buffer | |
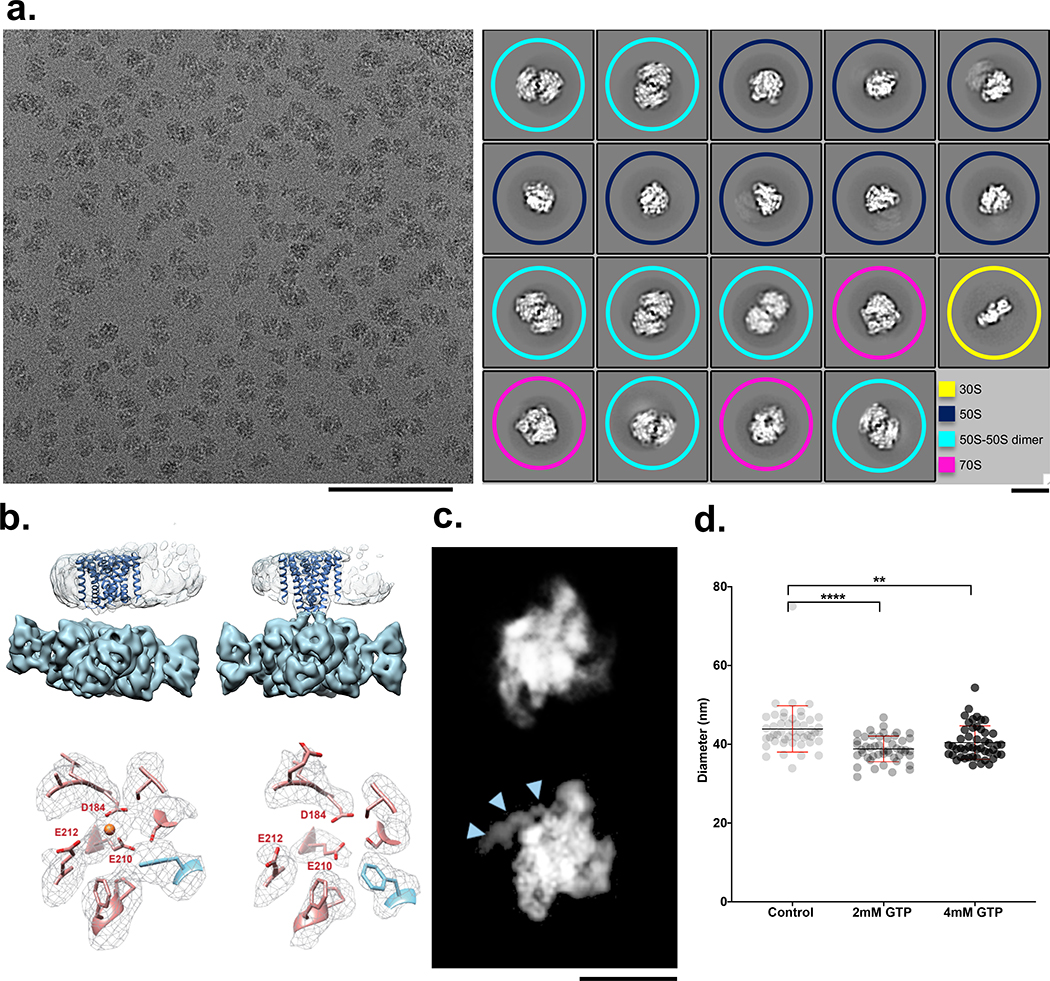
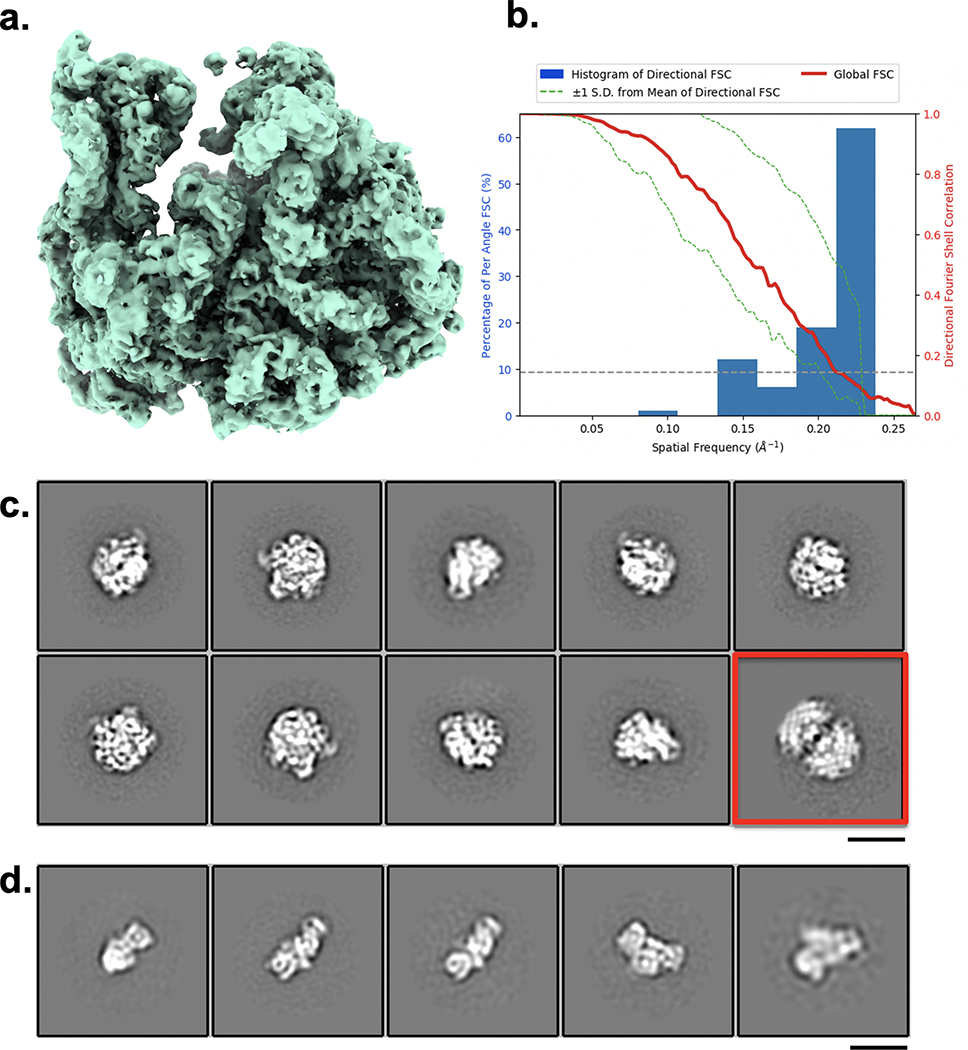
Early work on mixing-spraying demonstrated 30S and 50S ribosomal subunits associating into 70S ribosomes20. Figure 2a shows an example of an image of the overlapping area where these two samples have been mixed. Control experiments of the individual samples showed populations of 30S or 50S subunits (plus a small percentage of 50S dimer particles), and no evidence of 70S complexes (Extended Data Fig. 2c,d). The mixed sample contained 30S, 50S, 50S dimers and ~20% assembled 70S ribosomes (Fig. 2a); this particle count produced a reconstructed map of the 70S ribosome (FSC0.143, 4.8 Å) (Extended Data Fig. 2a,b). We note that previous work21 observed ~40% assembled 70S ribosomes using a mixing-spraying device followed by a 140 ms delay in a reaction chamber. This difference in assembly states is not surprising as the rate of interactions between subunits is expected to be much slower via diffusion within droplets that mix on the grid than by the mixing that occurs by chaotic advection in the device described in the earlier study. We also note that the previous study used a higher concentration of 30S subunits, twice that of 50S subunits, whereas the data shown here used a 1:1 ratio of 30S to 50S subunits.
Figure 2: Four examples of biological systems where time-resolved cryoEM provides answers.
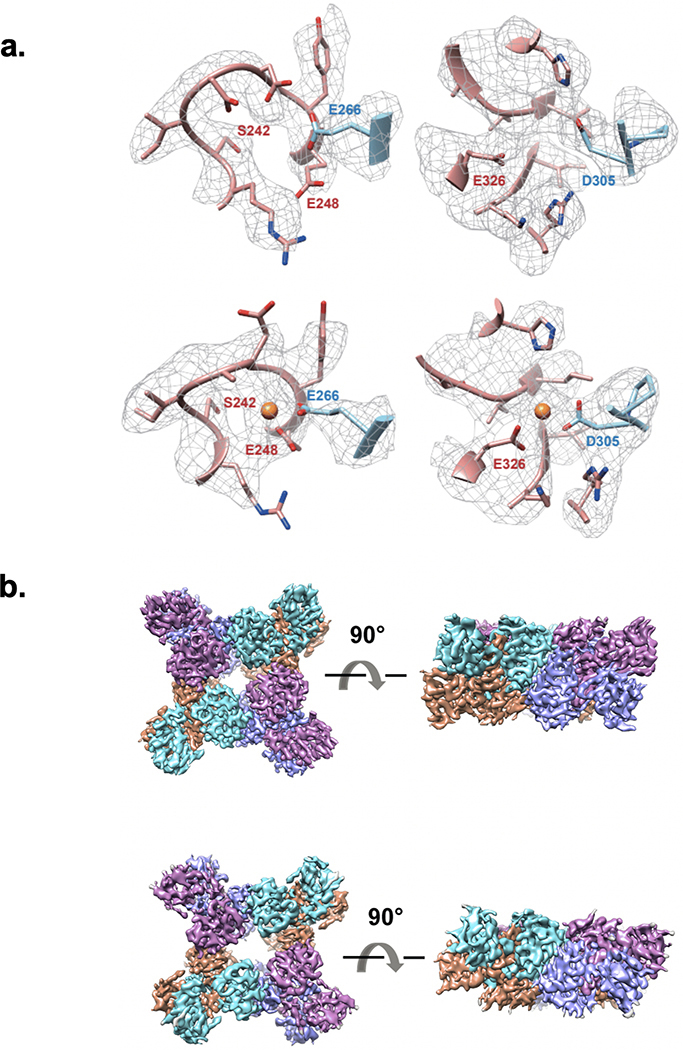
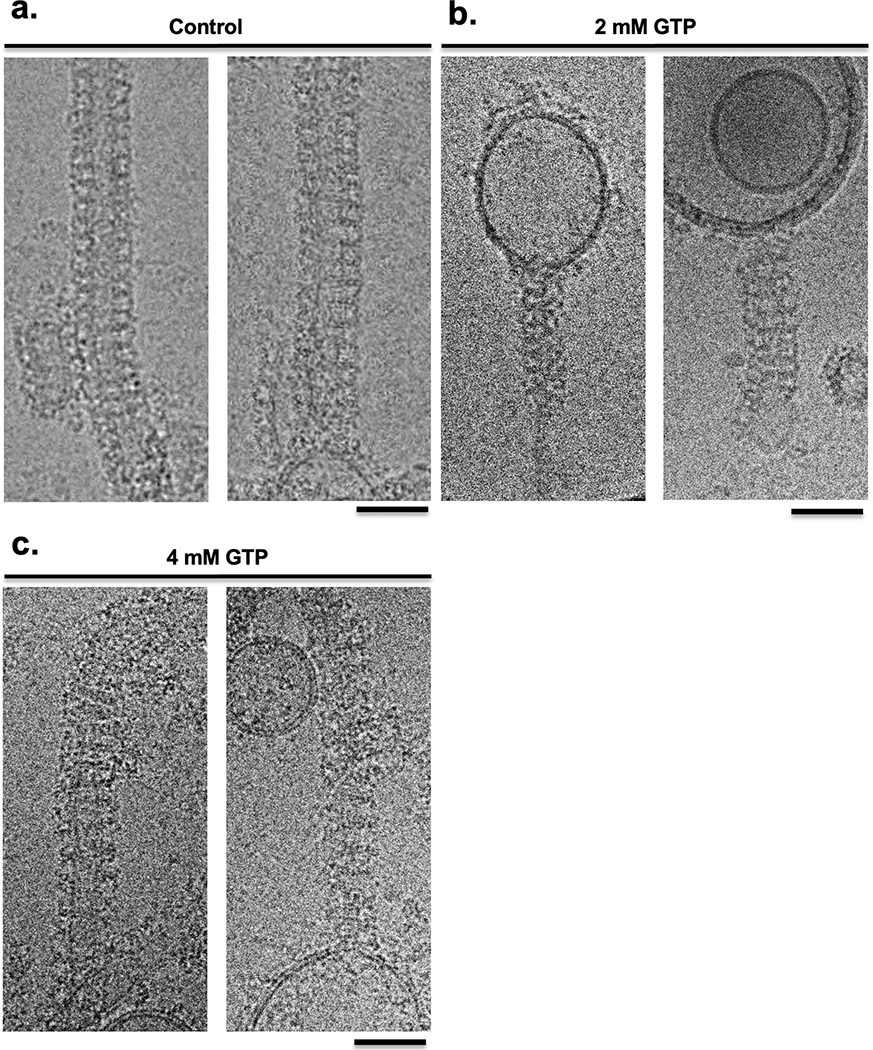
(a) left: Micrograph from the mixed region of a Spotiton prepared grid shows 30S and 50S ribosomal subunits and 70S complexes and is representative of 311 images. Scale bar, 100 nm; right: corresponding 2D classes show particles representing a population of 30S (yellow), 50S (blue), 50S-50S dimers (turquoise), and 70S (magenta). ~20% of particles were reconstructed to a 70S complex at a resolution of 4.75 Å (see Methods and Extended Data Fig. 2). Scale bar, 20nm. (b) 3D volumes generated from MthK in the presence (top left) or absence (top right) of calcium showing clear differences in the overall conformation of the channel. The bottom row shows one of the three Ca2+ binding sites in MthK either occupied in the case of a mixing experiment (left) or vacant as with the MthK only control (right). (c) Representative class averages of RNAP alone (top) or mixed with promoter DNA (bottom) showing DNA clearly bound (blue arrowheads). Scale bar, 10nm. (d) Measured diameters (mean ± SD) of dynamin-decorated tubes without (Control: 43.85, ± 5.86, n=48) and with GTP (2 mM: 38.80, ± 3.2, n=48; 4 mM: 40.40, ± 4.23, n=48,). **P = 0.0014, ****P = 0.0001, two-sided Student’s t test.
A compelling use of time-resolved cryoEM is observing changes in an ion channel during the early stages of its interaction with ligands. We examined conformational changes in a calcium-gated potassium channel (MthK22) upon adding Ca2+. MthK activates within milliseconds after Ca2+ application and inactivates slowly with a time constant of ~2 s23. This inactivation is caused by the 17 N-terminal residues obstructing permeation in a ball-and-chain-like manner 24. Single-particle cryoEM analyses of MthK in the absence and presence of Ca2+ yielded structures of closed and inactivated channels, respectively24. The prediction is that freezing MthK within ~100 ms of Ca2+ application, after the channel opens but before it inactivates, will reveal structures of open channels and other intermediates, completing the gating cycle. Using time-resolved Spotiton to mix MthK and Ca2+ at ~150 ms, the structure has the transmembrane domain tilted with respect to the ligand-binding domain (Fig. 2b, top). This tilt is an open-state signature and is different from the closed structure obtained without Ca2+ 24 (Fig. 2b, top). Analysis of the ligand-binding domain revealed that it corresponds to the open MthK gating ring structure24 (Extended Data Fig. 3b). In addition, densities for Ca2+ were observed at all known binding sites in the open MthK structure (Fig. 2b, bottom and Extended Data Fig. 3a). While the data is not yet sufficient to identify if the channel is open or inactivated - as the density for the transmembrane domains within the nanodiscs is weak - these results indicate that Ca2+ not only successfully mixed and bound to MthK, but also induced a conformational change to an activated state.
As another example of the use of Spotiton to capture biologically critical isomerizations, we investigated the initial steps of transcription initiation. Formation of the transcriptionally-competent “open” complex (RPo) is a dynamic, tightly regulated process requiring multiple steps. For E. coli RNA polymerase (RNAP), mechanistic studies demonstrate that intermediates on the pathway of DNA opening interconvert on the timescale of subseconds to seconds25,26. Although “kinetically-significant” intermediates in RPo formation were identified decades ago27, their transient nature has prevented atomic resolution structural characterization as they interconvert in time. Here we examined DNA opening by E. coli RNAP holoenzyme at the λPR promoter using time-resolved Spotiton. The kinetics of RPo formation at λPR have been extensively characterized, allowing predictions of intermediate populations as a function of time and solution conditions25. Within the ~150 ms time of mixing and freezing only the earliest intermediates are predicted to be present. The 2D class averages from this experiment show DNA bound to RNAP in a conformation consistent with promoter recognition (Fig. 2c). Future experiments that vary time, solution conditions, and promoter sequence combined with 3D classification strategies are anticipated to reveal the on-pathway nucleation and propagation of the transcription bubble.
Finally, we looked at the dynamics of dynamin at the ~150 ms timescale. During GTP hydrolysis, dynamin constricts and pinches off invaginating clathrin-coated vesicles. Previous results have shown dynamin constricts the membrane within seconds and during this process the helical parameters transform from a 1-start to a 2-start helix28,29. However, the rate and mechanism of how the dynamin polymer constricts and rearranges during GTP hydrolysis remain unknown. Using time-resolved Spotiton to mix pre-formed dynamin tubes with GTP, we observed that at ~150 ms, a high percentage of the dynamin decorated tubes were constricted (i.e. the lumen of the lipid bilayer was reduced) to 39 nm upon mixing with 2 mM GTP compared to 44 nm for untreated controls (Fig. 2d). Upon mixing with 4 mM GTP, the dynamin polymer becomes disordered, constricts, and disassembles from the lipid bilayer (Fig. 2d and Extended Data Fig. 4). This work provides the first clues to the initial steps that lead to dynamin-mediated membrane constriction and fission. We expect further analysis incorporating a decreasing range of GTP concentrations will trap the reaction at the slowest step, allowing changes in the dynamin organization during early fission events to be observed.
These four biological cases represent a range of short-lived molecular states of high interest and demonstrate that samples can be successfully mixed on a grid and rapidly vitrified within ~100 ms to trap intermediates. This method uses very small quantities of material and is applicable to mixing together any two, or potentially more, samples to allow the capture of short-lived molecular states that appear between 50–500 ms after an initial interaction.
Methods
Spotiton Instrument
The Spotiton system11,12 was upgraded with a second set of identical dispense head components. A second piezo driven electric tip was mounted next to the first head 4.5 mm from the first tip (Extended Data Fig. 1b). The 4.5 mm pitch allows the two tips to simultaneously aspirate sample from two adjacent holder tubes also mounted at a 4.5 mm pitch. The second tip includes a manual fine adjustment screw to allow precise alignment between the two tips in the direction perpendicular to the plunge axis motion.
The second piezo electric tip is driven by an independent electronic drive (DE03 controller) which was added to the system. The plunge axis outputs a series of electrical pulses while plunging (distance between pulses is a parameter set to 0.25 mm). The plunge axis pulse output is tied to the trigger input of both DE03 controllers. Each DE03 controller can be setup to start firing its respective tip after a unique number of pulses (configurable by the user) relating to the position of the plunge axis. The fluidics of the second piezo electric tip are attached to a second syringe pump which was added to the system. The syringe pumps allow precise independent sample aspiration and cleaning of the tips.
Time resolution:
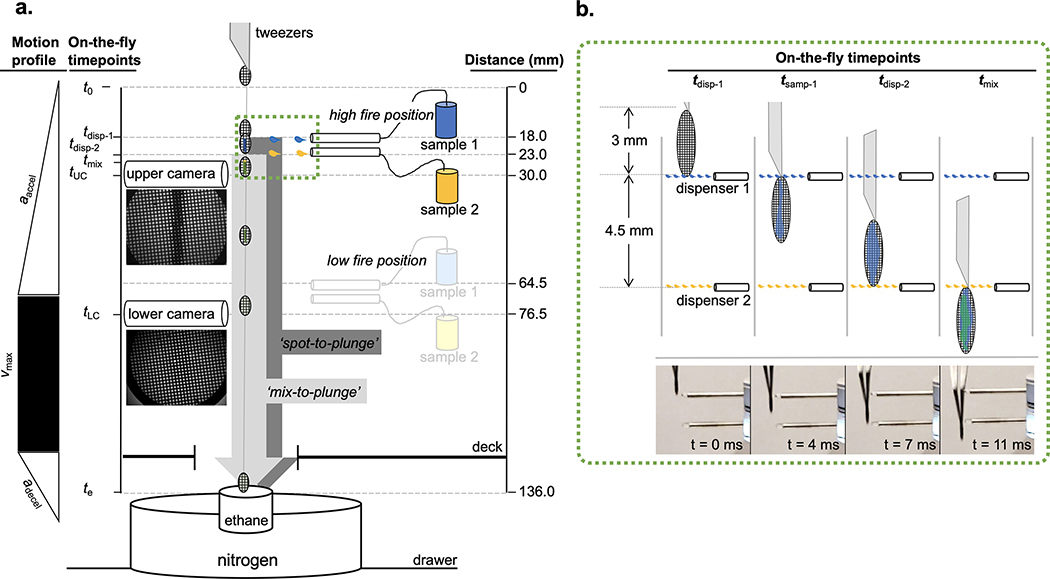
The motion path (136 mm) of a grid prepared by the Spotiton time-resolved system is characterized by three phases: acceleration, constant velocity, and deceleration (Extended Data Fig. 1a). The durations of the acceleration and constant velocity phases are variable and dependent on the rate of acceleration set by the user. The deceleration phase remains fixed and ends when the grid comes to a stop in the liquid ethane cup. The two dispensers spray a defined number of sample droplets whose first contact with the falling grid is separated by a period of time between 5–11 ms, depending on the acceleration rate selected (Supplementary Table 4). The first sample thus has a brief opportunity to be wicked away by the capillary action of the nanowires prior to contact by the second sample (Extended Data Fig. 1b). The mixing time of the two samples prior to vitrification ranges from 130–160 ms but can be reduced to ~90 ms by moving the dispensers to a “low-fire” position ~4 cm closer to the ethane bath (Condition 4 in Supplementary Table 1 and 2). A redesigned instrument would in principle be capable of even shorter mixing times; for example, the commercial version of the Spotiton system, chameleon (SPT Labtech), is capable of spot-to-plunge times of ~50 ms.
Time-resolved Spotiton operation:
A typical protocol for operating the time-resolved Spotiton system proceeds as follows. On startup, the system is initialized, and the two three-axis robots used to position the grid-holding tweezers and the dispensing heads are homed. Next, the fluid lines carrying distilled water from an external reservoir to the dispensing tips are flushed several times to remove air bubbles and any residual methanol used to clean the tips after the previous session. Both tips are then fired in view of an inspection camera to confirm successful dispensing and the formation of discrete droplets19. Next, a standard (not nanowire) test grid is loaded into the tweezers, lowered into position between the upper camera and the upper dispenser tip, and both are observed for alignment in the live viewer in the main software window. Aided by an integrated image-recognition algorithm, the operator positions the tip along the vertical midline and at the leading (lower) edge where the first dispensed droplets will contact the grid. The second piezo device is locked into position directly beneath the first, but its tip is steerable to allow the second sample to be dispensed either completely overlapping the first sample strip or in a discrete, non-contacting parallel strip. To verify operation, each tip is fired separately on the test grid and video captures from the upper camera are examined to confirm deposition of a liquid stripe. To verify tip alignment (i.e. both stripes are deposited onto the same grid area), both tips are fired simultaneously, and the video capture is examined for the presence of a single overlapping liquid stripe. Once successful two-tip dispensing on the test grid is confirmed, the humidity of the chamber is increased to 80–85% by activating the in-chamber nebulizer. Next, 5 μL of each sample is loaded into the sample holder cups, placed in the humidified chamber, and simultaneously aspirated into the two tips. Successful firing is again confirmed as described above.
The system is now ready to prepare vitrified grids. First, the upper tip is positioned in front of the upper camera as before and a plasma treated, nanowire grid is loaded into the tweezers. To avoid saturating the nanowires with water, thereby reducing the wicking capacity, exposure of the grid to the high humidity environment within the chamber is limited to 10–60 s prior to plunging, depending on the observed performance of the particular batch of nanowire grids being used. During a typical plunge, a grid acceleration of 10 m/s2 and a tip firing frequency of 14,750 Hz results in the deposition of ~70 droplets (~4 nl volume) of each sample onto the grid. This is observed as a single thick, opaque band of liquid down the grid in the upper camera video capture that wicks to a thin film that is nearly invisible in the lower camera video capture acquired 49 ms later, just before the grid plunges into the liquid ethane (Extended Data Figure 1a). To generate control grids with two non-overlapping sample strips separated by several squares, the lower, steerable tip can be adjusted to bring the tips out of alignment, as mentioned above, or more simply, we can change the acceleration of the grid. During our characterization of the system, we noted that when the tips are aligned to form completely overlapping stripes at a set acceleration (e.g. 10 m/s2), the stripes can, at other accelerations, become misaligned, e.g. merging of only the leading (8 m/s2; see Fig. 1a) or trailing (5 m/s2) ends, or even deposited as two parallel and completely separated stripes (6 m/s2). While we do not fully understand the mechanics of this phenomenon, it is reliably reproducible and can be used to make a control grid with two well separated streams of sample by a simple adjustment of the acceleration.
Compared to the original Spotiton protocols12, grid preparation and timing was adjusted to account for wicking of double the usual sample volume. This required optimizing our self-wicking grids13 to have longer length and higher density nanowires to create a faster and higher volume wicking area.
Sample preparation
A series of experiments was performed to first test and verify mixing of protein samples on the grids and then to demonstrate the value of this approach for a variety of biological systems of interest. In general, vitrified grids of mixed samples were prepared as follows. Nanowire grids were freshly plasma cleaned and transferred into the enclosed Spotiton chamber (room temperature, ~80–85% humidity) no more than 30 seconds prior to vitrification. 5 μL of each sample was loaded into the two sample holder cups with concentrations as tabulated in Table 1. For control experiments, the second sample was replaced by the carrying buffer as noted. The calculated spot-to-plunge time for all of the grids is 151 ms. Below we briefly describe further details of sample and grid preparation for each of these experiments.
Apoferritin and 70S ribosomes:
Apoferritin was purchased from Sigma Aldrich, 400 kDa, A3660, 2.3 mg/ml. Protein solution stored in 50% glycerol was exchanged into a cryo compatible buffer (50 mM Tris-Cl [pH 7.6]; 150 mM NaCl) using Amicon Ultra-15 centrifugal filter units (100 kDa cutoff membrane). 70S ribosomes were purchased from New England BioLabs Inc, 2MDa. Protein solution was stored in 20 mM HEPES-KOH [pH 7.6], 10 mM Mg(CH3COO)2, 30 mM KCl, and 7 mM β-mercaptoethanol after diluting the sample to 1 mg/ml from 33.3 mg/ml.
30S an 50S ribosomal subunits:
70S ribosomes are prepared as described in30. For subunit purification, 70S ribosomes were exchanged into dissociation buffer (20 mM MES-KOH [pH 6], 600 mM KCl, 8 mM Mg(CH3COO)2, 1 mg/ml heparin, 0.1 mM PMSF, 0.1 mM benzamidine, and 2 mM DTT) before loading onto a sucrose gradient in the same buffer and centrifuged for 19 hr at 28,500 RPM in the Ti25 rotor. The 50S and 30S subunits were exchanged separately into reassociation buffer (10 mM MES-KOH [pH 6], 10 mM NH4CH3COO, 40 mM CH3COOK, 8 mM Mg(CH3COO)2, and 2 mM DTT), concentrated to 6 μM, and stored at −80°C after being flash frozen in liquid nitrogen.
RNA Polymerase plus promoter DNA:
Core RNAP (subunit composition α2ββ’ω) was expressed and purified as described31. The specificity subunit σ70 was expressed and purified as described31 with the following modifications: i. a plasmid encoding His(6)-SUMO- σ70 was used instead of His(10)-SUMO- σ70; ii. cells were grown at 30°C in the presence of 50 μg/mL kanamycin until OD 0.4, then temperature was lowered to 16°C; iii. when the cells reached OD 0.7, 0.1 mM IPTG was added and growth continued for an additional 15 hrs. After harvest by centrifugation and resuspension in lysis buffer31, cells were flash frozen in liquid nitrogen and stored overnight at −80°C. Cells were thawed halfway at 22°C, thawed completely on ice, and then lysed in a French press. After lysis, the series of columns and buffers used to purify σ70 were as described31. For promoter DNA, a duplex λPR promoter fragment (−85 to +20) was used (Trilink Biotechnologies, San Diego, CA). Top (non-template) strand: ‘5 C GGA ATC GAG GGA TCC TCT AGA GTT GGA TAA ATA TCT AAC ACC GTG CGT GTT GAC TAT TTT ACC TCT GGC GGT GAT AAT GGT TGC ATG TAC TAA GGA GGT TGTA G 3’. Bottom (template-strand): 5’ C TACA ACC TCC TTA GTA CAT GCA ACC ATT ATC ACC GCC AGA GGT AAA ATA GTC AAC ACG CAC GGT GTT AGA TAT TTA TCC AAC TCT AGA GGA TCC CTC GAT TCC G 3’. RNAP holoenzyme was assembled by mixing core with a 3.3 molar excess of σ70, incubating for 20 min at 37°C, and buffer exchanging into gel filtration (GF) buffer (40 mM Tris-HCl [pH 8.0], 120 mM KCl, 10 mM MgCl2 and 10 mM DTT) using centrifugal filtration (Amicon-Ultra-0.5 m 30K cutoff) at 4°C. Excess σ70 was separated from core RNAP on a Superose 6 increase 10/300 GL column (GE Healthcare) equilibrated in GF buffer. The eluted fractions of RNAP were concentrated to 16 mg/ml (centrifugal filtration), aliquoted, flash frozen in liquid nitrogen, and stored at −80°C. The non-template and template strands of λPR promoter DNA were dissolved in annealing buffer (10 mM Tris-HCl [pH 8], 50 mM KCl, 0.1 mM EDTA), mixed in equimolar amounts and incubated in a 95°C heat block for 10 min. The samples were then slow cooled in the heat block to room temperature. Annealed DNA was stored at −80°C. RNAP and DNA aliquots were thawed on ice, diluted to the concentrations reported in Table 1 with GF buffer. N-octyl-β-D-glucopyranoside (Anatrace) was added to 0.35% final just before spraying at room temperature.
Ca2+ activated channel MthK:
MthK was purified and reconstituted into nanodiscs composed of 3:1 POPE:POPG, following the procedure described in detail previously23,24.
Dynamin tubes plus GTP:
Liposome formation and dynamin purification: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (100 μl of 5 mg/ml, DOPS, Avanti) was dried and resuspended in 250 μl HCB150 (50 mM HEPES, 150 mM KCl, 2 mM EGTA, 1 mM MgCl2, 1 mM TCEP, [pH 7.5]). Unilamellar liposomes were obtained by extruding the mixture 21 times through a 0.4 μm pore-size polycarbonate membrane (Avanti). Recombinant ΔPRD-dynamin 1 was purified from Sf9 insect cells. Briefly, recombinant baculovirus containing the sequence of ΔPRD-dynamin 1 with an N-terminal His-tag, was generated by following Bac-to-Bac Baculovirus Expression System (ThermoFisher Scientific). The suspension cultures of Sf9 were maintained in Sf-900 III serum-free media (SFM, ThermoFisher Scientific) and inoculated with recombinant baculovirus at a cell density of 1.6 × 106 with 1/100 volume of virus/final volume of medium. The cells were grown for 72 h at 27°C, and pelleted by centrifugation at 1000 x g, 5 min, 4°C. The pellet was resuspended in 3 μl modified HSB150 (50 mM HEPES, 150 mM KCl, 5 mM beta-mercaptoethanol,10 mM imidazole, [pH 8.0]) and containing protease inhibitor cocktail (Millipore Sigma). The cells were then lysed by sonication (total time of 8 min with 5 s pulse-on and 15 s pulse-off) followed by high speed centrifugation (20,000 x g, 15 min). The supernatant was collected, passed through Ni-NTA beads and the protein was eluted with 150 mM imidazole in modified HSB150. The protein solution was dialyzed in HSB150 overnight and the purity was checked using SDS-PAGE/Coomassie staining. Dynamin decorated tubes were generated by incubating 3 μl of DOPS liposomes with 40 μl of protein (0.8 mg/ml, in 10 mM Tris, 10 mM KCl, 1 mM MgCl2, [pH 7.4]) for 2 h.
Imaging and analysis
Typically, data was acquired using Leginon MS32 and micrographs were collected either on a Titan Krios (Thermo Fisher Scientific) with a K2 or K3 BioQuantum counting camera (Gatan, Inc.) operating in counting mode or on a Tecnai F20 equipped with a TVIPS CMOS camera. The nominal magnification, pixel size, exposure time, frame rate, total dose, and defocus range were as shown in Table 2 for each experiment. For all datasets, frames were aligned using MotionCorr233 and CTF was estimated using Ctffind434.
Table 2:
CryoEM imaging parameters
| Sample | Microscope/ Detector | Nominal magnification | Pixel size | Dose rate (e−/px/sec) | Total dose (e−/Å2) | Frame rate (ms) | Defocus range (um) | Total images | Particles selected |
|---|---|---|---|---|---|---|---|---|---|
| 30S + 50S mixed | Titan Krios/K2 | 105,000 | 1.096 | 8 | 64.61 | 200 | 1.4 – 3 | 311 | 18,434 |
| 50S only | Titan Krios/K2 | 130,000 | 0.855 | 8 | 69.13 | 200 | 1.5 – 3 | 3586 | 604,271 |
| 30S only | Titan Krios/K2 | 130,000 | 0.855 | 8 | 69.13 | 200 | 1.5 – 3 | 668 | 25,412 |
| MthK + Ca2+ mixed | Titan Krios/K3 | 81,000 | 1.0825 | 4 | 42.18 | 50 | 1 – 3 | 4329 | 1,310,918 |
| MthK only | Titan Krios/K2 | 81,000 | 1.0825 | 8 | 43.57 | 50 | 1 – 3 | 7420 | 956,882 |
| RNAP + DNA mixed | Titan Krios /K2 | 29,000 | 0.832 | 8 | 69.74 | 200 | 1.2 – 2.2 | 3535 | 745,167 |
| RNAP only | Titan Krios/K3 | 10,500 | 1.096 | 4 | 67.26 | 200 | 1.2 – 2.2 | 3945 | 277,685 |
| Dynamin + GTP Mixed | F20/K2 | 29,000 | 1.27 | 8 | 40 | 250 | 1.5 – 3 | 74 | - |
| Dynamin only | F20/TVIPS | 62,000 | 3 | - | 62.17 | - | 2.5 | 100 | - |
70S association complex, 50S and 30S ribosomal subunits:
Particle picking was performed with Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) and extracted in Relion by 5X binning followed by one round of 2D classification using Relion35 to remove false particles. After this first round of 2D classification, classes clearly representing 50S dimers and 30S and 50S particles (Fig. 2a) were excluded from further steps of image analysis. After one round of 3D classification, only recognizable 70S particles were selected and reextracted to a pixel size of 2.2 Å for final refinement. A total of 26,402 particles were used for homogeneous 3D refinement in Relion resulting in a 4.8 Å map of the 70S ribosome (Extended Data Fig. 2a,b). For the control experiments, a procedure similar to that described above was used to obtain a total of 12,505 and 3,762 individual 50S and 30S particles, respectively, and 2D classified in Relion (Extended Data Fig. 2c,d).
MthK with and without Ca2+:
Typically, a small set of particles was manually picked, and 2D class averages were calculated using the CL2D algorithm36 inside the Appion image processing pipeline37. A subset of these classes was used as templates to pick particles for the entire set of micrographs using FindEM38. For MthK without Ca2+, from 956,882 particles and after several rounds of 2D and ab initio classification in Cryosparc39. 428,917 particles were used for a final 3D classification and the best class was selected and used for Cryosparc2 non-uniform refinement to generate a structure with an overall resolution of 4.2 Å as shown in top right of Figure 2b.
For MthK with Ca2+, a procedure was used similar to that described above. Briefly, 2,158,345 particles were auto picked and used for 2 rounds of 2D classification in Relion3. From these, 849,864 good particles were selected and used for 3D classification in Relion3. The open state class with a highly tilted RCK domain was selected and used for Cryosparc2 non-uniform refinement to generate a structure with an overall resolution of 6.3 Å as shown in top left of Figure 2b.
For the focused refinement of the RCK domain, signal subtracted particles of both samples were generated with a mask to only include the RCK domain. These particles were used for refinement in Relion3 applying C2 symmetry and the overall resolution is 4.1 Å for Mthk with Ca2+ and 3.5 Å for MthK without Ca2+ (Extended Data Fig. 3b).
RNAP with and without DNA:
Particle picking was performed with Gautomatch and extracted in Relion followed by one round of 2D classification to remove false picks using 2D classification tool in Cryosparc39. 167,212 particles of RNAP with λPR promoter DNA and 52,747 particles of RNAP alone are used for another round of 2D classification and the 2D classes with high resolution features were selected (Figure 2c).
Dynamin with and without GTP:
46, 28, and 100 images were collected for dynamin-decorated tubes mixed with 4 mM GTP, 2 mM GTP and no GTP, respectively. For each condition, the diameters of 48 tubes were measured using Fiji40.
Extended Data
Extended Data Fig. 1.
Specifications of time-resolved Spotiton operation
(a) Diagrammatic overview of the distances (fixed) and elapsed times (variable) relevant to spraying and mixing two samples on a moving grid. Simultaneous dispensing of both samples is triggered after the grid plunge begins. Representative images from the upper and lower cameras are shown directly below the illustrations of each. Sample 1 and sample 2 are indicated in blue and yellow, respectively. (b) Magnified view of (green-dashed) boxed area in (a) showing grid and dispensing at specific time-points with corresponding high-speed video captures of the tips and grid below. Elapsed times shown on each image reflect estimates from a video of a grid plunged under Condition 2 (Supplementary Table 1). Objects in (a) and (b) are not drawn to scale. Supplementary Tables 1–4 list values for the following parameters of a grid plunged as depicted in (a) and (b): aaccel, acceleration rate; adecel, deceleration rate; vmax, maximum velocity; t0, plunge start point; tdisp-1, grid leading edge reaches first dispenser; tsamp-1, sample 1 fully applied to grid; tdisp-2, grid leading edge reaches second dispenser; tmix, samples 1 and 2 fully applied to grid; tUC, grid reaches upper camera, tLC, grid reaches lower camera; te, grid plunges into ethane. ‘Spot-to-plunge’ and ‘mix-to-plunge’ in (a) reflect the elapsed times from tdisp-1 or tmix to te, respectively.
Extended Data Fig. 2.
Mixing 30S and 50S ribosomal subunits to form 70S complexes
(a) ~20% of particles present were reconstructed to 70S complex at a resolution of 4.75 Å as indicated by FSC0.5 (b). (c) 2D classes of 50S ribosomal subunit obtained from the control experiment; 2D class of 50S-50S dimer is shown in red. (d) 2D classes of the 30S ribosomal subunit obtained from the control experiment. Both control experiments show no evidence of 70S ribosomes as observed in the mixed experiment. Scale bars, 20 nm.
Extended Data Fig. 3.
Cryo-EM maps of MthK RCK domain with and without Ca2+
(a) The two additional Ca2+ binding sites of MthK either vacant from a control experiment (top row) or occupied after mixing with calcium (bottom row). (b) 3D volumes of MthK RCK domains without (top row) and with (bottom row) Ca2+ bound.
Extended Data Fig. 4.
Mixing of GTP with dynamin-decorated lipid tubes results in constriction
Representative cryo-electron micrographs of control dynamin-decorated tubes without GTP (a), with 2 mM GTP (b) and 4mM GTP (c). Scale bars, 50 nm.
Supplementary Material
Acknowledgements
We are grateful to the staff of the Simons Electron Microscopy Center at the New York Structural Biology Center for help and technical support. We thank I. Fernandez and B. Huang for kindly providing the ribosome subunits. We thank H. He (NIDDK, NIH) for cryo-EM data collection, NIDDK EM Core Facility, and the Hinshaw lab for critical comments. The work presented here was conducted at the National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the NIH (GM103310, OD019994) and the Simons Foundation (SF349247) and the NIDDK NIH Intramural Research Program. Other grant support includes NIH/NIGMS R35 GM118130 (to S.A.D) and NIH/RO1GM088352 (to C.M.N.).
Footnotes
Ethics (Competing Interests) Declaration
B.C./C.S.P. have a commercial relationship with SPT Labtech, a company that produces a commercially available instrument, chameleon, that is based on the Spotiton prototype.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Frank J Time-resolved cryo-electron microscopy: Recent progress. J Struct Biol 200, 303–306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulder AM et al. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330, 673–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriman J & Unwin N Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 56, 241–252 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Unwin N Acetylcholine receptor channel imaged in the open state. Nature 373, 37–43 (1995). [DOI] [PubMed] [Google Scholar]
- 5.White HD, Walker ML & Trinick J A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. J Struct Biol 121, 306–313 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Unwin N & Fujiyoshi Y Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol 422, 617–634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramaniam S et al. Protein conformational changes in the bacteriorhodopsin photocycle. J Mol Biol 287, 145–161 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Lu Z et al. Gas-Assisted Annular Microsprayer for Sample Preparation for Time-Resolved Cryo-Electron Microscopy. J Micromech Microeng 24, 115001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Z et al. Passive Microfluidic device for Sub Millisecond Mixing. Sens Actuators B Chem 144, 301–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Z et al. Key Intermediates in Ribosome Recycling Visualized by Time-Resolved Cryoelectron Microscopy. Structure 24, 2092–2101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain T, Sheehan P, Crum J, Carragher B & Potter CS Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J Struct Biol 179, 68–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandey VP et al. Spotiton: New features and applications. J Struct Biol 202, 161–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei H et al. Optimizing ‘self-wicking’ nanowire grids. J Struct Biol 202, 170–174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scapin G et al. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 556, 122–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z et al. Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han H et al. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y et al. FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noble AJ et al. Routine single particle CryoEM sample and grid characterization by tomography. eLife 7, e34257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu JLY, Tellkamp F, Khajehpour M, Robertson WD & Miller RJD Rapid mixing of colliding picoliter liquid droplets delivered through-space from piezoelectric-actuated pipettes characterized by time-resolved fluorescence monitoring. Rev Sci Instrum 90, 055109 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Lu Z et al. Monolithic microfluidic mixing-spraying devices for time-resolved cryo-electron microscopy. J Struct Biol 168, 388–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B et al. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure 23, 1097–1105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zadek B & Nimigean CM Calcium-dependent gating of MthK, a prokaryotic potassium channel. J Gen Physiol 127, 673–685 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posson DJ, Rusinova R, Andersen OS & Nimigean CM Calcium ions open a selectivity filter gate during activation of the MthK potassium channel. Nat Commun 6, 8342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C et al. Ball-and-chain inactivation in a calcium-gated potassium channel. Nature (2020) doi: 10.1038/s41586-020-2116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruff EF, Record MTJ & Artsimovitch I Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazumder A & Kapanidis AN Recent Advances in Understanding sigma70-Dependent Transcription Initiation Mechanisms. J Mol Biol 431, 3947–3959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saecker RM, Record MTJ & Dehaseth PL Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412, 754–771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundborger AC et al. A dynamin mutant defines a superconstricted prefission state. Cell Rep 8, 734–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L et al. Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 560, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references
- 30.Johansson M, Bouakaz E, Lovmar M & Ehrenberg M The kinetics of ribosomal peptidyl transfer revisited. Mol Cell 30, 589–598 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Chen J et al. E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorzano COS et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171, 197–206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lander GC et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roseman AM FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol 145, 91–99 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.