Abstract
The Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has resulted in outbreak of global pandemic, fatal pneumonia in human referred as Coronavirus Disease-2019 (Covid-19). Ayurveda, the age old practice of treating human ailments in India, can be considered against SARS-CoV-2. Attempt was made to provide preliminary evidences for interaction of 35 phytochemicals from two plants (Phyllanthus amarus and Andrographis paniculata used in Ayurveda) with SARS-CoV-2 proteins (open & closed state S protein, 3CLpro, PLpro and RdRp) through in silico docking analysis. The nucleotide analogue remdesivir, being used in treatment of SARS-CoV-2, was used as a positive control. The results revealed that 18 phytochemicals from P. amarus and 14 phytochemicals from A. paniculata shown binding energy affinity/dock score < − 6.0 kcal/mol, which is considered as minimum threshold for any compound to be used for drug development. Phytochemicals used for docking studies in the current study from P. amarus and A. paniculata showed binding affinity up to − 9.10 kcal/mol and − 10.60 kcal/mol, respectively. There was no significant difference in the binding affinities of these compounds with closed and open state S protein. Further, flavonoids (astragalin, kaempferol, quercetin, quercetin-3-O-glucoside and quercetin) and tannins (corilagin, furosin and geraniin) present in P. amarus have shown more binding affinity (up to − 10.60 kcal/mol) than remdesivir (up to − 9.50 kcal/mol). The pharmacokinetic predictions suggest that compounds from the two plants species studied in the current study are found to be non-carcinogenic, water soluble and biologically safe. The phytochemicals present in the extracts of P. amarus and A. paniculata might have synergistic effect with action on multiple target sites of SARS-CoV-2. The information generated here might serve as preliminary evidence for anti SARS-CoV-2 activity of phytochemicals present from P. amarus and A. paniculata and the potential of Ayurveda medicine in combating the virus.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-020-02578-7.
Keywords: Andrographis paniculata, Coronavirus, COVID-19, Ligands, Molecular docking, Natural and Phyllanthus amarus
Introduction
The outbreak of highly transmissible fatal pneumonia in human referred as Coronavirus Disease-2019 (Covid-19) has spread globally (215 countries and territories) with over 16.81 M confirmed cases and over 662,095 deaths worldwide as on 31 July 2020 (https://www.worldometers.info/coronavirus), whereas, in India, 15, 83,792 people with 34,968 deaths (https://www.worldometers.info/coronavirus). According to the International Committee on Taxonomy of Viruses (ICTV), the novel zoonotic pathogenic virus associated with COVID-19 is Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the virus species is severe acute respiratory syndrome-related coronavirus (Gorbalenya et al. 2020). The break out of SARS-CoV-2 has been first reported during December 2019 in Wuhan, Hubei Province, China, with a fatality rate of 4 percent (Liu et al. 2020; Wu et al. 2020; Zhu et al. 2020). Sore throat, dry cough, fever, chills, repeated shaking with chills, muscle pain, headache, loss of taste or smell and respiratory distress are major symptoms induced by SARS-CoV-2 (Cheng et al. 2007). In most severe cases, the clinical presentation of SARS-CoV-2 infection includes pneumonia that progresses to multi-organ failure and even death (Zaki et al. 2012; Machhi et al. 2020). Covid-19 has become global pandemic and created the wide spread fear of death everywhere in the world resulting lockdown in human society (Mandal 2020). The increasing pandemic potential of SARS-CoV-2 is due to its high human-to-human transmissible efficiency which makes it difficult to contain (Wu et al. 2020; Zhao et al. 2020).
The SARS-CoV-2 is a β-coronavirus, belongs to family Coronaviridae of order Nidovirales (Zhu et al. 2020). It has enveloped positive sense single-stranded RNA genome with 33 kilo base (Kb) encoding for 14 open reading frames (ORFs) (Fehr and Perlman 2015; Guo et al. 2020). The genome of SARS-CoV-2 lacks the hemagglutinin-esterase gene. However, it comprises two flanking untranslated regions (UTRs) at 5′ end and 3′ with 265 and 358 nucleotides, respectively. The first ORF comprises approximately 67 percent of the genome and encodes 16 non-structural proteins (nsps), which were processed by proteolytic cleavage, whereas the remaining 13 ORFs encode for structural proteins and protein accessory (Boopathi et al. 2020; Chan et al. 2020). The nsps mainly include helicase, RNA-dependent RNA polymerase (RdRp), 3-Chymotrypsin-like protease (3CLpro), Papain-like protease (PLpro) and others likely to be involved in the transcription and replication of SARS-CoV-2 (Simmons et al. 2005; Boopathi et al. 2020). Four major structural proteins are spike surface glycoprotein (S protein), membrane protein (M), nucleocapsid protein (N) and envelope (E). M protein possess N-terminal glycosylated ectodomain at the N-terminal end that consists of three transmembrane domains (TM) and a long C-terminal CT domain (Boopathi et al. 2020; Chan et al. 2020). The M and E proteins are required for virus morphogenesis, assembly and budding, whereas S protein is a fusion viral protein comprising two subunits S1 and S2. The S1 subunit consists of receptor-binding domain (RBD), N-terminal domain (NTD) and signal peptide. The S2 subunit comprises two heptad repeat regions known as HR-N and HR-C, which form the coiled-coil structures surrounded by the protein ectodomain (Walls et al. 2020). A furin cleavage site (PRRARS’V) is present in S protein at the interface between S1 and S2 subunits that will be processed during the biogenesis (Coutard et al. 2020). SARS-CoV-2 S protein facilitates the entry of viral particles into the host cell after binding with the host angiotensin-converting enzyme 2 (ACE-2) (Tortorici and Veesler 2019) and also determines the host range and cellular tropism (Simmons et al. 2005). Entry of SARS-CoV-2 into host cells is mediated by the cleavage of viral spike protein by host proteases, viz. serine proteases (TMPRSS2) or cysteine proteases (Cathepsin B/L) (Boopathi et al. 2020; Yan et al. 2020). The ACE2 and TMPRSS2 proteins are prime targets of the coronavirus (Matsuyama et al. 2010; Glowacka et al. 2011; Zhao et al. 2020).
The highly conserved spike protein (S protein) of SARS-CoV-2 enters human cells by targeting the host ACE2. Hence, S protein is one of the drug target to neutralize the virus (Walker and Burton 2018; Karakus et al. 2020). Proteases (3CLpro and PLpro) translated from viral RNA are essential for polyprotein processing of SARS-CoV-2 and involved in replication and transcription of SARS-CoV-2 can serve as potent therapeutic targets (Prussia et al. 2011; Sheahan et al. 2020). The SARS-CoV-2 proteases act on polyproteins (PP1a and PP1ab) to release the nsps which are encoded by open reading frame (ORF1a/b) of virus (Sheahan et al. 2020). Inhibiting virus entry into the cell and its replication combined with modulating the immune system could be a potential target for drug therapy (Sanders et al. 2020). Currently, there are no available vaccines or specific medicines for the treatment of COVID-19.
Several countries and pharma giants in the world are working to come out with either vaccines or drugs which will be helpful in containing this global pandemic (Begum et al. 2020). In India, attempts were made to address this problem through Ayurveda and Siddha practices as well apart from development of drugs and vaccines, which were still widely used among the Indian population to combat diseases caused by viral infections such as jaundice, malaria, cholera, etc. (Gomathi et al. 2020). Ayurveda is the age old practice of treating human ailments in India and provided remedies against many medical conditions. Medicinal herbs from India are promising in treating various illnesses (Gomathi et al. 2020). Further, identification of phytochemicals from medicinal plants can be used as drugs for viral infection targeting viral receptors (Chang and Woo 2003; Keyaerts et al. 2004), viral integration (Kim et al. 2010), reverse transcription, viral replication and viral protein translation (Mansouri et al. 2009).
Extracts from Andrographis paniculata Wall (Kalmegh) and Phyllanthus amarus Schum. & Thonn were used in Ayurvedic medicinal systems and proven to have numerous medicinal properties (Niranjan et al. 2010; Patel et al. 2011). A. paniculata also known as ‘King of bitters’ belonging to Acanthaceae family (Pholphana et al. 2013) is widely used to treat fever, diarrhoea and common cold. It consists of a wide range of pharmacologically active substances such as andrographolides and its derivatives that have been showed anticancer, antimalarial, hepatoprotective, antioxidant activities (Niranjan et al. 2010) and reported to have potent antiviral activity against diverse group of viruses belonging to different families (Gupta et al. 2017). It also been implicated in suppression of increased NOD-like receptor protein 3 (NLRP3), caspase-1 and interleukin-1β molecules which are extensively involved in the pathogenesis of SARS-COV and likely SARS-CoV-2 as well (Liu et al. 2020a, b). Phyllanthus amarus Schum. & Thonn. (Family: Euphorbiaceae) is one of the medicinal herb widely distributed in tropical and subtropical countries from Africa to Asia, South America and the West Indies. It is a rich source of secondary metabolites such as alkaloids, flavonoids, hydrolysable tannins, lignins, polyphenols, triterpenes, sterols and volatile oils (Patel et al. 2011). It exhibits an array of ethanopharmacological activities such as anti-inflammatory, hepatoprotective, nephroprotective, anti-amnesia, anticancer, diuretic, antioxidant, antiviral, anti-bacterial, anti-hyperglycemic, anti-hypercholesterolemia and so on (Calixto et al. 1998; Patel et al. 2011).
Several Ayurvedic practitioners in India claiming Ayurveda medicine can cure Covid-19, which are not supported by enough scientific evidences. Nevertheless, the role of herbal extracts in addressing the human ailments cannot be ignored totally. Lack of proper experimental evidences might have made regulatory authorities to enforce not to sale the Ayurveda medicines as a curative agent of Covid-19. There are several Ayurveda tablets available in the Indian market claiming to boost immune system in the backdrop of Covid-19 outbreak. Here we discussed on the potential Ayurveda medicine prepared using A. paniculata and P. amarus against SARS-CoV-2 through the results of in silico analysis of interaction between SARS-CoV-2 proteins and secondary metabolites of these plants available in the databases. This provides some preliminary scientific evidence to harness the potential of Ayurveda medicines in combating SARS-CoV-2. However, this needs to be to be validated with experimental evidences required by the regulatory authorities such as large-scale clinical trials.
Materials and methods
Sources of ligands and viral receptor proteins
The list of phytochemicals present in P. amarus and A. paniculata was obtained from previously published scientific literature (Chao and Lin 2010; Gupta and Vaghela 2019; Joseph and Raj 2011; Nisar et al. 2018; Pereira et al. 2016; Tan et al. 2016). A total of 35 compounds were selected for the molecular docking studies (21 from P. amarus and 14 from A. paniculata) (Table 1). Remdesivir, an antagonist of SARS-CoV-2 RdRp, was chosen as a positive control (Yu et al. 2020).
Table 1.
Dock score of interaction between phytochemicals from Phyllanthus amarus and Andrographis paniculata with SARS-CoV-2 proteins; S protein [PDB ID:6VSB,6VXX (closed), 6YVB (open)], 3 chymotrypsin like protease (3CLpro) (PDB ID:6LU7), papain like protease (PLpro) (PDB ID:4OVZ) and RNA-dependent RNA polymerase (RdRp) (PDB ID:6NUS)
| Group | Sl. No. | Compound | S protein | S protein (Closed) | S protein (Open) | 3CLpro | PLpro | RdRp |
|---|---|---|---|---|---|---|---|---|
| Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | Binding affinity (kcal/mol) | |||
| Positive control | 1 | Remdesivir | − 8.00 | − 8.40 | − 8.50 | − 7.90 | − 9.50 | − 7.60 |
| Phytochemicals from P.amarus | ||||||||
| Alkaloids | 1 | Epibubbialine | − 6.80 | − 7.30 | − 6.70 | − 5.90 | − 7.50 | − 6.30 |
| 2 | Isobubbialine | − 7.40 | − 8.20 | − 7.30 | − 6.00 | − 7.70 | − 6.40 | |
| 3 | nor-securinine | − 6.80 | − 7.20 | − 7.20 | − 5.70 | − 7.30 | − 6.80 | |
| 4 | Securinine | − 7.10 | − 7.40 | − 7.10 | − 5.90 | − 7.70 | − 7.30 | |
| Flavonoids | 5 | Astragalin* | − 9.80 | − 8.30 | − 8.30 | − 8.40 | − 9.70 | − 7.60 |
| 6 | Kaempferol* | − 8.30 | − 8.30 | − 8.40 | − 7.70 | − 9.60 | − 7.00 | |
| 7 | Quercetin* | − 8.10 | − 8.80 | − 8.80 | − 7.50 | − 8.60 | − 8.60 | |
| 8 | Quercetin-3-O-glucoside* | − 9.80 | − 8.60 | − 8.60 | − 8.20 | − 10.30 | − 7.70 | |
| 9 | Quercitrin* | − 9.20 | − 8.60 | − 8.60 | − 8.60 | − 8.80 | − 8.10 | |
| Phenylpropanoid | 10 | Cinnamic acid | − 5.80 | − 5.70 | − 6.00 | − 4.70 | − 6.30 | − 5.60 |
| 11 | Hinokinin* | − 8.40 | − 8.50 | − 8.40 | − 7.40 | − 9.80 | − 8.20 | |
| 12 | Niranthin | − 6.30 | − 5.90 | − 6.10 | − 6.30 | − 7.60 | − 6.10 | |
| 13 | Nirtetralin | − 7.40 | − 7.00 | − 7.00 | − 6.60 | − 6.70 | − 7.00 | |
| 14 | Phenylalanine | − 5.50 | − 5.70 | − 5.80 | − 4.80 | − 6.00 | − 5.60 | |
| 15 | Phyllanthin | − 6.10 | − 5.60 | − 5.90 | − 5.70 | − 7.60 | − 6.40 | |
| Tannins | 16 | Corilagin* | − 8.80 | − 9.60 | − 9.50 | − 8.70 | − 8.50 | − 9.30 |
| 17 | Furosin* | − 10.20 | − 9.60 | − 9.40 | − 8.70 | − 9.20 | − 9.10 | |
| 18 | Geraniin* | − 10.60 | − 10.10 | − 10.10 | − 9.30 | − 9.20 | − 9.10 | |
| 19 | Melatonin | − 6.60 | − 6.60 | − 7.20 | − 5.80 | − 7.00 | − 5.50 | |
| Triterpenes | 20 | farnesyl farnesol | − 5.20 | − 5.40 | − 5.80 | − 4.50 | − 5.80 | − 4.70 |
| 21 | Phenazine | − 6.70 | − 7.00 | − 7.10 | − 5.80 | − 7.50 | − 7.00 | |
| Phytochemicals from A. paniculata | ||||||||
| Bioactive compounds | 1 | Andrograpanin | − 7.30 | − 7.10 | − 6.60 | − 6.10 | − 6.80 | − 7.20 |
| 2 | Andrographolide | − 7.90 | − 7.50 | − 7.60 | − 6.50 | − 8.00 | − 7.30 | |
| 3 | Isoandrographolide* | − 9.10 | − 7.30 | − 8.10 | − 7.00 | − 9.30 | − 8.30 | |
| Diterpenes | 4 | 14-acetylandrographolide | − 7.60 | − 7.50 | − 8.00 | − 6.70 | − 8.40 | − 7.00 |
| 5 | 14-deoxy-11,12-didehydroandrographolide | − 7.70 | − 7.60 | − 7.70 | − 7.60 | − 7.30 | − 7.10 | |
| 6 | 14-deoxy-14,15-didehydroandrographolide | − 8.00 | − 8.20 | − 7.40 | − 6.80 | − 7.80 | − 7.90 | |
| 7 | 14-Deoxyandrographolide | − 8.00 | − 8.20 | − 7.60 | − 7.30 | − 8.30 | − 7.80 | |
| 8 | 19-O-acetyl-14-deoxy-11,12-didehydroandrographolide | − 8.40 | − 7.40 | − 7.50 | − 6.80 | − 8.80 | 7.10 | |
| 9 | Deoxyandrographolide | − 7.80 | − 7.40 | − 8.10 | − 7.30 | − 7.00 | 7.50 | |
| 10 | Neoandrographolide | − 8.20 | − 8.10 | − 8.40 | − 7.80 | − 7.80 | − 8.50 | |
| Flavonoids | 11 | 5-hydroxy-7,8,2′,3′-tetramethoxyflavone, | − 7.60 | − 8.00 | − 7.40 | − 6.90 | − 8.40 | − 7.00 |
| 12 | 5-hydroxy-7,8,2′,5′-tetramethoxy flavone | − 8.20 | − 7.90 | − 7.50 | − 7.60 | − 8.60 | − 7.80 | |
| 13 | 5-hydroxy-7,8-dimethoxyflavanone | − 7.80 | − 7.40 | − 7.80 | − 7.20 | − 8.90 | − 7.30 | |
| 14 | 5-Hydroxy-7,8-dimethoxyflavone | − 7.90 | − 7.20 | − 8.00 | − 7.50 | − 9.00 | − 7.50 | |
The docking score of respective phytochemicals higher than the positive control (remdesivir) has been denoted in bold
*The phytochemicals showing better binding affinity with all the viral proteins
The three-dimensional (3D) conformers of the selected ligands were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov) database in SDF and MOL formats. Similarly, the 3D structures of proteins determined using a biophysical technique such as X-ray crystallography or NMR spectroscopy are openly available in protein data bank were retrieved (www.rcsb.org). Four different viral targets were considered for docking studies, viz. spike protein (S protein), 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro) and RNA-dependent RNA polymerase (RdRp). The crystal structure of S protein and 3CLpro of SARS-CoV-2 available in the protein databank were retrieved for the current study. The PDB ID for S protein and 3CLpro are PDB ID: 6VSB, chains A, B, C, 6VXX (closed), 6VYB (open) (Wrapp et al. 2020) and PDB ID: 6LU7, chain A (Jin et al. 2020), respectively. Crystal protein structures of PLpro and RdRp of SARS-CoV-2 were not available in the protein databank at the time of analysis. Hence, the crystal structure developed for the both proteins using the X-ray structure of PLpro of human coronavirus papain-like proteases (PDB ID: 4OVZ, chain A) (Baez-Santos et al. 2014) and the solved RdRp structure (97.08% sequence identity with SARS-CoV-2) of SARS-HCoV (PDB ID: 6NUS, chain A) (Kirchdoerfer et al. 2019) were downloaded for docking studies.
Preparation of ligands and target proteins
The retrieved 3D structures of the ligands were optimized using Avogadro version 1.2.0 (Hanwell et al. 2012) with force filed type MMFF94 and converted to PDB format using Open Babel version 3.1.1. (O’Boyle et al. 2011). For further simplification of analysis, the optimized ligands with the lowest energy were loaded to AutoDock-MGL Tools (Morris et al. 2009), the Gasteiger charges were added and standard processes were used to obtain the PDBQT files. The downloaded 3D structures were checked for the presence of any improper bonds, side-chain anomalies and missing hydrogens using PyMOL software (Delano 2009). All the water molecules, complex molecules, ions and ligands of the proteins were removed in Biovia Discovery Studio 2020 (Biovia 2020). The optimized PDB structures were uploaded to AutoDock-MGL Tool (Morris et al. 2009), succinctly, polar hydrogens were added and standard processes were used to obtain the PDBQT files.
Active site prediction and molecular docking
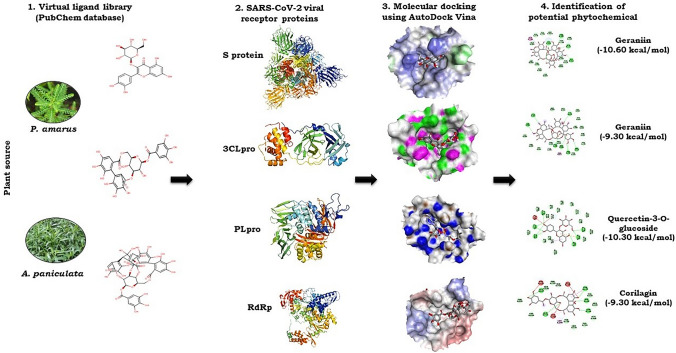
The active sites of the proteins were determined using the Computed Atlas for Surface Topography of Proteins (CASTp) (http://sts.bioe.uic.edu/castp/index.html?201l) and Biovia Discovery Studio 2020. For closed and open conformational state of spike protein, the active sites were chosen based on the receptor-binding domain, which interacts with the ACE2 receptors on the host cell membrane of humans (Gur et al. 2020). In case of 3CLpro protein, the binding site was selected based on the location of co-crystallized inhibitor N3, which was developed using computer-aided drug design and has shown specific inhibition to SARS and MERS with good binding affinity in the binding pockets of crystal structure of 3CLpro (Jin et al. 2020). The optimized ligands and proteins in PDBQT format were subjected to molecular docking with the aid of AutoDock Vina software (Trott and Olson 2010). AutoDock Vina predicts the interaction between protein and ligand by utilizing its scoring function (binding affinity). In the process of molecular docking, based on predicted active sites, the grid box was set to 60 Å × 60 Å × 60 Å centred at 225.616, 226.490, 225.337 (XYZ coordinates) for spike protein 6VSB, grid box was set to 80 Å × 80 Å × 80 Å centred at 223.220962, 190.082990, 263.907088 (XYZ coordinates) for spike proteins 6VXX (closed) and 6VYB (open), grid box 26 Å × 26 Å × 26 Å centred at − 10.712, 12.411, 68.8312 (XYZ coordinates) for 3CLpro, grid box 60 Å × 60 Å × 60 Å centred at − 8.611, 38.916, − 41.012 (XYZ coordinates) for PLpro and grid box 60 Å × 60 Å × 60 Å centred at 138.751, 163.504, 136.636 (XYZ coordinates) for RdRp. The value of exhaustiveness was set at 24 (Froli et al. 2016). The output file obtained after docking analysis consists of top nine binding poses with their respective binding affinity in kcal/mol. The ligand binding pose showing highest binding affinity with least root mean square deviation (RMSD) was selected. The protein–ligand interaction in 3D structures was visualized in PyMOL. Similarly, 2D structure was visualized in Biovia Discovery studio 2020. The 3D visualization represents the exact location or the binding pocket of the target protein, whereas the 2D structure visualization represents the different bonds formed between the amino acid residues of the viral target protein and the ligand. The workflow of in silico molecular docking analysis is represented in Fig. 1.
Fig. 1.
Workflow of molecular docking analysis of phytochemicals present in P. amarus and A. paniculata with four proteins (S protein, 3CLpro, PLpro and RdRp) targets of SARS-CoV-2. The phytochemicals of the two plant species were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov) database in SDF and MOL formats. The 3D structures of proteins available in protein data bank were retrieved (www.rcsb.org). The detailed protocol is given in the material and methods
ADME and other pharmacokinetic properties
The drug-likeness analysis was carried out based on Lipinski’s rule of five (Lipinski 2004). Absorption, distribution, metabolism and excretion (ADME) are pharmacokinetic properties of a ligand that deals primarily with its absorption, distribution, metabolism and excretion in the human body. The major parameter, Lipinski’s rule of five was generated using SWISSADME (Daina et al. 2017) for evaluating the pharmacological significance. The ligands in PDB format were converted to canonical SMILES format and uploaded to SWISSADME prediction (http://www.swissadme.ch). Properties such as solubility (logS), human intestinal absorption (HIA) and carcinogen were obtained from admetSAR (http://lmmd.ecust.edu.cn/admetsar2).
Results and discussion
Impact of Covid-19 is having catastrophic effect on human health and life. The scientific communities from the nuke and corner of the world have scrupulously engaged in developing the drug or vaccine against SARS-CoV-2, causal agent of Covid-19. The current study discusses on the potential of phytochemicals present in two medicinal plants used in the Ayurveda, an age old medical practice in India. Plants/herbal extracts part of Ayurveda medicines are known to boost immunity in humans, which is crucial for fighting against microbial enemies (Kumar et al. 2020). Plant extracts from P. amarus and A. paniculata have been extensively used in Ayurveda for curing various human ailments (Niranjan et al. 2010; Patel et al. 2011). The herbal extracts may have several phytochemicals, few of them might be playing role in neutralizing the pathogens. Binding interaction between a pathogen protein and ligand derived from herbal extracts may result in inhibition of the activity of pathogen protein. Hence, to understand and determine the potency of the plant extracts against cataclysmic virus SARS-CoV-2, about 21 compounds present in P. amarus and 14 compounds present in A. paniculata were used for molecular docking simulation with four SARS-CoV-2 receptor targets.
Prior to the docking analysis, the ligands were optimized by minimizing the energy with force field type MMFF94, this helps in removing clashes among atoms and develops a stable starting pose of the ligands for binding interaction (Klebe et al. 2006). The docking coupled with a scoring function can be utilized to evaluate a large number of potential drugs in silico. In the process of molecular docking, generally, the binding affinity lesser than the upper threshold (− 6 kcal/mol) is regarded as a cut off value for determining good binding affinity between ligand and protein (Shityakov et al. 2014). Of the 35 compounds screened from two plant species (Table 1), three compounds (cinnamic acid, phenylalanine and farnesyl farnesol) did not qualify as per the threshold binding affinity to neutralize SARS-CoV-2 proteins.
In the recent clinical trials, remdesivir, a nucleotide analogue of adenosine triphosphate has been reported to be a potential prodrug against SARS-CoV-2 infection (Gao et al. 2020). It was originally developed for treating viral infection caused by Ebola and Marburg virus but was also found effective against SARS and MERS (Elfiky 2020). Hence, remdesivir has been used as a positive control in the current study. During virus replication, the RdRp mistakenly incorporates remdesivir into its genome, which results in stalling of replication (Wang et al. 2020). In our study, remdesivir has shown good interaction with RdRp protein (Table 1) with the binding affinity of − 7.6 kcal/mol. Similarly, it has shown a good binding affinity with 3CLpro (− 7.90 kcal/mol), S protein (− 8.00 kcal/mol) and PLpro (− 9.50 kcal/mol) (Table 1) and might be having some role in inhibiting SARS-CoV-2. Many recent computational investigations also showed inhibition activity of remdesivir with various viral receptors of SARS-CoV-2 (Elfiky 2020; Hall and Feng 2020; Murugan et al. 2020). This shows that the prodrug might be effective against all the four viral receptors.
The 3D and 2D visualization of top three phytochemicals along with positive control remdesivir based on binding affinity with respective viral receptor protein is represented in Figs. 2, 3, respectively. Hydrogen bond energy is the major contributor in dock score. Hydrogen bond formation between ligand structures and viral receptors is responsible for inhibition of the target protein, and it reflects the firmness of bonding between the protein and ligand (Chen et al. 2016). Selected phytochemicals from P. amarus and A. paniculata used in the current docking studies against SARS-CoV-2 proteins shown very good dock score above the threshold cut-off of − 6 kcal/mol. Ligand structures and necessary hydrogen bond formation between phytochemical with respective viral receptor is illustrated in Table 2 and supplementary Fig. 1.
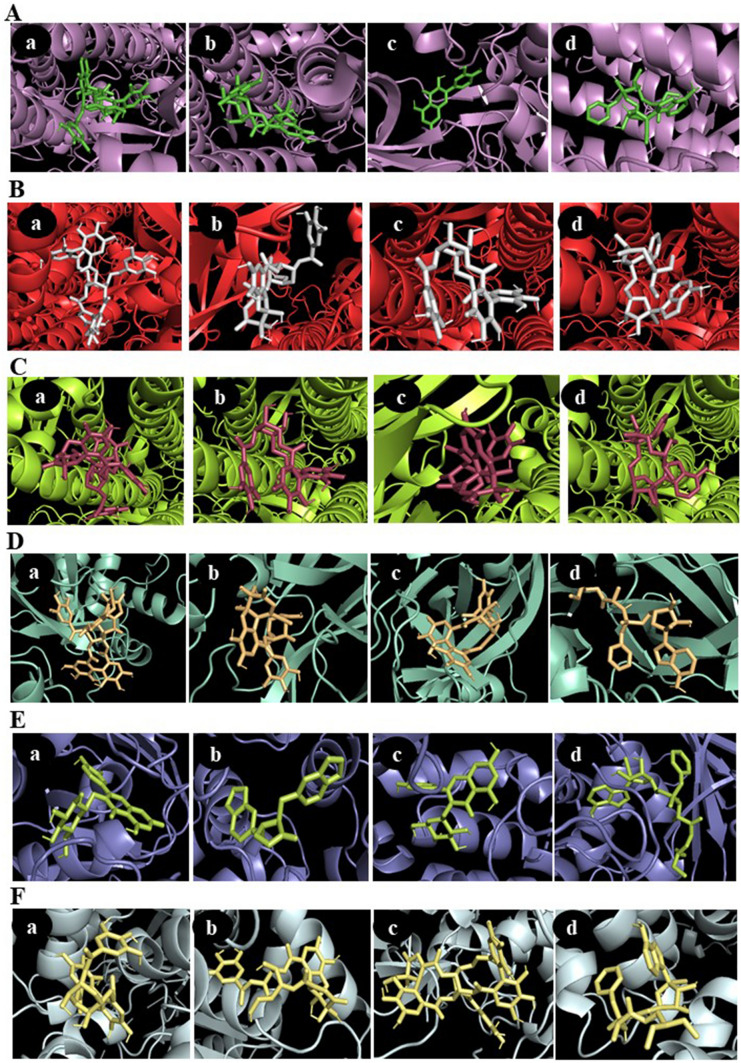
Fig. 2.
Output file obtained after docking analysis consists of top 9 binding poses with their respective binding affinity in kcal/mol. The ligand-binding pose showing highest binding affinity with least root mean square deviation (RMSD) was selected. The protein–ligand interaction in 3D structures were visualized in PyMOL. 3D visualization of interaction between SARS-CoV-2 proteins with representative phytochemicals. A S protein (6VSB) with a geraniin, b furosin, c quercetin-3-O-glucoside and d remdesivir (positive control). B S protein closed (6VXX) with a geraniin, b furosin, c corilagin and d Remdesivir (positive control) C S protein closed (6VYB) with a geraniin, b corilagin, c furosin and d remdesivir (positive control). D 3CLpro (6LU7) with a geraniin, b furosin, c corilagin and d remdesivir (positive control). E PLpro (4OVZ) with a quercetin-3-O-glucoside, b hinokinin, c astragalin and d remdesivir (positive control). F RdRp (6NUS) with a corilagin, b furosin, c geraniin and d remdesivir (positive control)
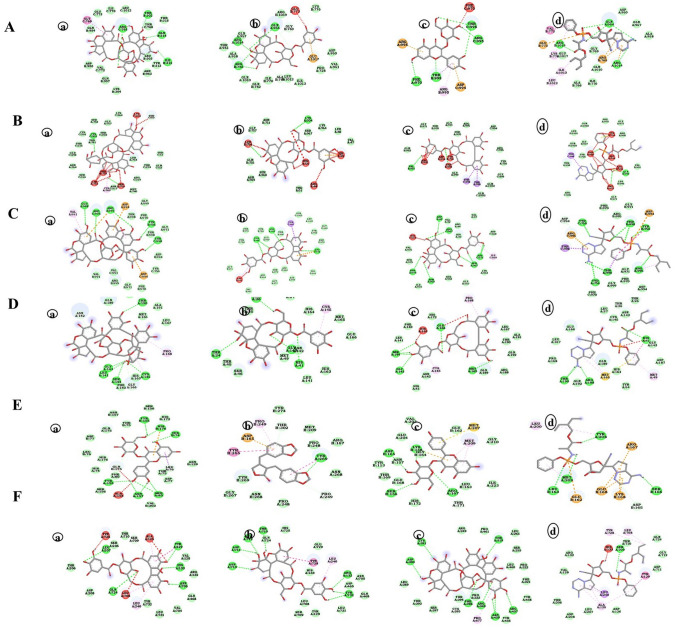
Fig. 3.
Hydrogen bond energy is the major contributor in dock score. Hydrogen bond formation between ligand structures and viral receptors is responsible for inhibition of the target protein and it reflects the firmness of bonding between the protein and ligand. After docking analysis, the interaction of protein receptor and ligand was visualized in 2D with the aid of discovery studio software. The figure represents 2D visualization of interaction between SARS-CoV-2 proteins with representative phytochemicals. The various bonds formed between the amino acid residues of SARS-CoV-2 proteins and ligands have been depicted. A S protein (6VSB) with a geraniin, b furosin, c quercetin-3-O-glucoside and d remdesivir (positive control). B S protein closed (6VXX) with a geraniin, b furosin, c corilagin and d remdesivir (positive control). C S protein closed (6VYB) with a geraniin, b corilagin, c furosin and d remdesivir (positive control). D 3CLpro (6LU7) with a geraniin, b furosin, c corilagin and d remdesivir (positive control). E PLpro (4OVZ) with a quercetin-3-O-glucoside, b hinokinin, c astragalin and d remdesivir (positive control). F RdRp (6NUS) with a corilagin, b furosin, c geraniin and d remdesivir (positive control)
Table 2.
Number of hydrogen bonds formed during the interaction between phytochemical structures with SARS-CoV-2 Proteins; S protein [PDB ID:6VSB,6VXX (closed), 6YVB (open)], 3 chymotrypsin like protease (3CLpro) (PDB ID:6LU7), papain like protease (PLpro) (PDB ID:4OVZ) and RNA-dependent RNA polymerase (RdRp) (PDB ID:6NUS). The interactions showing more binding affinity than remdesivir and SARS-CoV-2 are presented here
| Sl. No. | Compound with PubChem ID | Structural and chemical formula | No. of H bonds | Proteins of SARS-CoV-2 | Amino acid residues of viral receptor involved in hydrogen bonding with ligand |
|---|---|---|---|---|---|
| 1 |
Remdesivir 121,304,016 |
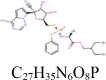
|
5 | S protein | GLN954, ARG1014, ARG1019 |
| 4 | 3CLpro | HIS41, AGR188, THR190 | |||
| 4 | PLpro | SER156, LEU163, TYR208, MET209 | |||
| 1 | RdRp | SER709 | |||
| 2 |
Astragalin 5,282,102 |
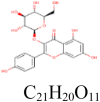
|
3 | S protein | PHE970, ASP994, THR998 |
| 2 | 3CLpro | SER144, ARG188 | |||
| 4 | PLpro | SER156, LYS158, ASP165, ARG167 | |||
| 3 |
Kaempferol 5,280,863 |
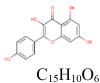
|
4 | S protein | TYR756, ARG995, THR756 |
| 5 | PLpro | THR75, ASP77, ARG83, ASN157 | |||
| 4 |
Quercetin 5,280,343 |
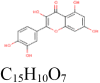
|
4 | S protein | THR549, ASP745, ASN978, ARG1000 |
| 2 | S protein (Closed) | TYR741 | |||
| 10 | S protein (Open) | TR549, ASN978, GLY744, ARG1000, THR573, MET740, TYR741 | |||
| 4 | RdRp | GLU350, PRO323, ASN628, VAL675 | |||
| 5 |
quercetin-3-O-glucoside 5,280,804 |
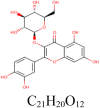
|
5 | S protein | PHE970, ARG995, THR998 |
| 1 | S protein (Closed) | THR998 | |||
| 4 | S protein (Open) | THR998, ARG995, ASP994 | |||
| 3 | 3CLpro | LEU141, HIS163, MET 165 | |||
| 8 | PLpro | HIS74, ARG83, TYR155,ASN157,HIS176 | |||
| 3 | RdRp | ALA125, HIS133 | |||
| 6 |
Quercitrin 5,280,459 |
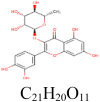
|
8 | S protein | ASP994, AGR995, PHE970, THR998 |
| 1 | S protein (Closed) | THR998 | |||
| 3 | S protein (Open) | ASP994, VAL991 | |||
| 1 | 3CLpro | ASN142 | |||
| 1 | RdRp | TYR732 | |||
| 7 |
Hinokinin 442,879 |
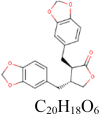
|
4 | S protein | THR573, LEU977,ASN978, AGR1000 |
| 0 | RdRp | ||||
| 1 | PLpro | TYR269 | |||
| 8 |
Corilagin 73,568 |
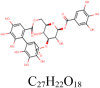
|
5 | S protein | GLU773, LYS947, GLU1017, ASN1023 |
| 2 | S protein (Closed) | VAL991, THR998 | |||
| 4 | S protein (Open) | THR998, AGR995 | |||
| 6 | 3CLpro | GLY143, SER144, MET165, GLU166 | |||
| 5 | 6NUS | TYR129, HIS133, LEU207, ASN705, GLN724 | |||
| 9 |
Furosin 10,416,810 |
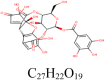
|
4 | S protein | ARG765, GLN954, AGG1014 |
| 2 | S protein (Closed) | LYS304, LEU754 | |||
| 7 | S protein (Open) | ASP40, ARG567, ASP571, SER967 | |||
| 4 | 3CLpro | THR24, THR26, ASN142, GLY143 | |||
| 6 | PLpro | ASP165, PRO249, GLN270, TYR274 | |||
| 7 | RdRp | ARG132, THR710, ASP711, GLY712, ASN713, TYR732 | |||
| 10 |
Geraniin 3,001,497 |
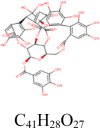
|
4 | S protein | THR302, ILE312, GLN314, ARG765 |
| 2 | S protein (Closed) | CYS291, SER750 | |||
| 5 | S protein (Open) | GLU988, ARG995, TYR756, THR998 | |||
| 5 | 3CLpro | LEU141, GLY143, SER144, CYS145, THR190 | |||
| 2 | PLpro | GLU251, TYR269 | |||
| 8 | RdRp | THR319, ASP390, LYS391, ARG349, PHE396, ARG457, ASN459 | |||
| 11 |
Isoandrographolide 343,585 |
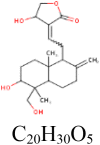
|
2 | S protein | GLU773, ARG1019 |
| 1 | PLpro | TYR155 | |||
| 0 | RdRp | ||||
| 12 |
14-deoxy-14,15-didehydroandrographolide 6,473,762 |
|
1 | RdRp | SER236 |
| 13 |
14-Deoxyandrographolide 11,624,161 |
|
1 | RdRp | SER236 |
| 14 |
19-O-Acetyl-14-deoxy-11,12-didehydroandrographolide 46,179,874 |
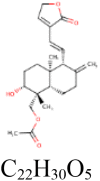
|
4 | S protein | ARG319, LEU977, ARG1000 |
| 15 |
Neoandrographolide 9,848,024 |
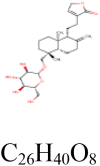
|
2 | S protein | GLU748, ASN856 |
| 4 | RdRp | ALA125, ASP126, ARG132, HIS133 | |||
| 16 |
5-hydroxy-7,8,2′,5′-tetramethoxy flavone 16,215,023 |
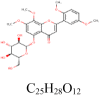
|
4 | S protein | GLN314, GLN613, ILE666, LYS733 |
| 2 | RdRp | GLN728, TYR728 |
Interaction between spike glycoprotein (S protein) [6VSB, 6VXX (closed), 6VYB (open)] and phytochemicals
S protein of the SARS-CoV-2 latches onto human angiotensin-converting enzyme 2 (ACE2) receptor and undergoes structural changes that allow the viral membrane to fuse with the cell membrane of the host (Walls et al. 2020). Despite the high similarity between sequences and structures of SARS and SARS CoV-2, the drugs successfully used to cure SARS are ineffective in case of this novel virus (Fani et al. 2020). This indicates that more number of potential molecules should be screened to find an effective molecule.
The molecular docking analysis of S protein of the SARS-CoV-2 with 21 selected phytochemicals from P. amarus showed that the majority of the compounds bind to S protein with favourable binding energy ranging from − 6.10 kcal/mol (for phyllanthin) to − 10.60 kcal/mol (for geraniin) (Table 1). When the binding energy of phytochemicals was investigated, nine compounds, viz. geraniin (− 10.60 kcal/mol), furosin (− 10.20 kcal/mol), astragalin (− 9.80 kcal/mol), quercetin-3-O-glucoside (− 9.80 kcal/mol), quercitrin (− 9.20 kcal/mol), corilagin (− 8.80 kcal/mol), hinokinin (− 8.40 kcal/mol), kaempferol (− 8.30 kcal/mol) and quercetin (− 8.10 kcal/mol), exhibited highest binding affinity. Interestingly, the phytochemicals belonging to the flavonoids and tannins of P. amarus have shown a very high affinity with the target protein. Molecular dynamic simulation studies also reported that quercetin and kaempferol bind with the ACE2-S complex with low binding free energy (Pandey et al. 2020). The antiviral bioactivities of many flavonoids from medicinal plants were reported by several studies (Thayil et al. 2016; Zakaryan et al. 2017; Jo et al. 2020). Among all compounds, geraniin exhibited (− 10.60 kcal/mol) highest docking score with S protein. THR302, ILE312, GLN314 and ARG765 amino acid residues were involved in forming four hydrogen bonds in the binding pocket of S protein (Table 2, Fig. 3).
In binding interaction between phytochemical compounds (14) from A. paniculata with S protein of the SARS-CoV-2, the docking score ranged from − 7.30 (for andrograpanin) to − 9.10 kcal/mol (for isoandrographolide) (Table 1). Of the 14 compounds, five compounds: isoandrographolide (− 9.10 kcal/mol), 19-O-acetyl-14-deoxy-11, 12-didehydroandrographolide (− 8.40 kcal/mol), neoandrographolide (− 8.20 kcal/mol) and 5-hydroxy-7, 8, 2′, 5′-tetramethoxy flavone (− 8.20 kcal/mol) shown better binding affinity as compared to remdesivir (− 8.00 kcal/mol). Isoandrographolide has properly positioned into the binding pocket constructed by hydrogen bonding with GLU773 and ARG1019 amino acids of S protein exhibited highest (− 9.10 kcal/mol) binding affinity (Table 2, Fig. 2). Totally, 13 compounds from P. amarus and A. paniculata have shown better inhibitory activity against S protein.
The S protein undergoes conformational changes to produce closed and open state based on down and up positions, respectively, in the receptor-binding domain. Hence, docking analysis was carried out in both the conformational states with 21 selected phytochemicals from P. amarus and 14 phytochemicals from A. paniculata. There was no significant difference in the binding affinities of the compounds with both closed and open state S protein. The range of binding affinities of 21 selected phytochemicals from P. amarus with closed state S protein of SARS-CoV-2 was − 6.60 (for melatonin) to − 10.10 kcal/mol (for geraniin). Seven compounds, geraniin (− 10.10 kcal/mol), furosin (− 9.60 kcal/mol), corilagin (− 9.60 kcal/mol), quercetin (− 8.80 kcal/mol), quercetin-3-O-glucoside (− 8.60 kcal/mol), quercitrin (− 8.60 kcal/mol) and hinokinin (− 8.50 kcal/mol) have shown better binding affinity than remdesivir (− 8.40 kcal/mol). Among these, geraniin with maximum binding affinity of − 10.10 kcal/mol has formed two hydrogen bonds with CYS291, SER750 amino acid residues of closed state S protein. In case of open state S protein, the dock score of 21 phytochemicals from P. amarus ranged between − 6.10 kcal/mol (for niranthin) and − 10.10 kcal/mol (for geraniin). Six compounds, geraniin (− 10.10 kcal/mol), corilagin (− 9.50 kcal/mol), furosin (− 9.40 kcal/mol), quercetin (− 8.80 kcal/mol), quercetin-3-O-glucoside (− 8.60 kcal/mol) and quercitrin (− 8.60 kcal/mol) have shown favourable binding affinity than remdesivir (− 8.50 kcal/mol). Geraniin with maximum dock score of − 10.10 kcal/mol form five hydrogen bonds with amino acid residues, GLU988, ARG995, TYR756, THR998 of closed state S protein.
All the 15 phytochemicals of A. paniculata docked with both closed and open state S protein falls between − 6.00 kcal/mol and − 8.4 kcal/mol for closed state and − 8.50 kcal/mol (remdesivir). However, these phytochemicals might also have neutralizing effects on the activity of S protein since their dock score is more than − 6.00 kcal/mol, which is a cut-off value for considering the effective binding affinity. The binding affinities ranged from − 7.10 kcal/mol (for andrograpanin) to − 8.20 kcal/mol (for14-deoxy-14,15-didehydroandrographolide and 14-deoxyandrographolide) for closed state of S protein. For the open state S protein, dock score ranged between − 6.60 kcal/mol (for andrograpanin) and − 8.40 kcal/mol (for neoandrographolide). The results indicate that the phytochemicals under study do not have much significant difference with respect to binding affinities in both the closed and open state of S protein. However, there was difference observed with respect to the hydrogen bonding between open and closed state S protein with more favourable bonds observed in open state compared to closed state, which is having more unfavourable bonds.
Interaction between 3-chymotrypsin like protease (3CLpro) (6LU7) and phytochemicals
The SARS-CoV-2 protease enzyme, 3CLpro, involved in C-terminal cleavage of virus polyprotein is a potential target for drug discovery to SARS-CoV-2, since it controls replication (Qmar et al. 2020). Hence, this potential target was evaluated with the selected 35 compounds from P. amarus and A. paniculata. Before docking simulation, the target site for viral receptor protein was selected based on the location of co-crystallized inhibitor N3 as this target site has been considered to be the most commending site for ligand interaction (Murugan et al. 2020).
The binding affinity of compounds to 3CLpro ranged from − 6.10 kcal/mol (for andrograpanin) to − 9.30 kcal/mol (for geraniin) (Table 1). Among 21 phytochemicals derived from P. amarus, six compounds, viz. geraniin (− 9.30 kcal/mol), furosin (− 8.70 kcal/mol), corilagin (− 8.70 kcal/mol), quercitrin (− 8.60 kcal/mol), astragalin (− 8.40 kcal/mol) and quercetin-3-O-glucoside (− 8.20 kcal/mol), have shown better binding affinity as compared to remdesivir (− 7.90 kcal/mol) (Table 1). The compounds belong to tannins (geraniin, furosin and corilagin) and flavonoids (quercitrin, astragalin and quercetin-3-O-glucoside) are found to have good inhibitory action on 3CLpro. Several reports have been shown antiviral effect of flavonoids (Gescher et al. 2011; Chen et al. 2012). Recently, Adem et al. (2020) evaluated 80 flavonoids compounds present in different medicinal plants against 3CLpro of SARS-CoV-2. Among them, hesperidin, rutin, diosmin, apiin, diacetylcurcumin, (E)-1-(2-Hydroxy-4-methoxyphenyl)-3-[3-[(E)-3-(2-hydroxy-4- methoxyphenyl)-3-oxoprop-1-enyl] phenyl] prop-2-en-1-one and beta, beta'-(4-Methoxy-1,3- phenylene) bis(2′-hydroxy-4′,6′-dimethoxyacrylophenone have shown effective binding affinity with SARS-CoV-2 3Clpro. The flavonoids from the two plant species used in the present study have higher binding affinity to protease similar results were reported by Adem et al. (2020). Geraniin, with the highest binding affinity of − 9.30 kcal/mol, has formed five hydrogen bonds with the amino acid residues, LEU141, GLY143, SER144, CYS145 and THR190 of 3CLpro (Table 2, Fig. 3).
The docking score for compounds present in A. paniculata ranged from − 6.10 kcal/mol (for andrograpanin) to − 7.80 kcal/mol (for neoandrographolide) (Table 1) which is on par with the remdesivir showing the score of − 7.90 kcal/mol. During replication of the viral genome, the 3CLpro cleaves the C-terminal of polypeptide protein at 11 distinct sites to generate various nsps (Qmar et al. 2020). Hence, the phytochemicals present in the extracts of P. amarus might hamper the activity of the protease enzyme there by inhibiting the process of virus multiplication.
Interaction between papain like protease (PLpro) (4OVZ) and phytochemicals
The SARS-CoV-2 another protease enzyme, PLpro known to be involved in cleaving of N-terminal of viral polyprotein which is also an important antiviral drug target. It also plays a crucial role in its replication along with 3CLpro (Baez-Santos et al. 2014). The 3D structure of PLpro used in the docking simulation is the x-ray structure of SARS-CoV, which has the highest similarity with the PLpro of SARS-CoV-2.
The binding energy of phytochemicals present in P. amarus ranged from -6.30 kcal/mol (for cinnamic acid) to − 10.30 kcal/mol (for quercetin-3-O-glucoside) (Table 1). Four compounds; quercetin-3-O-glucoside (− 10.30 kcal/mol), hinokinin (− 9.80 kcal/mol), astragalin (− 9.70 kcal/mol) and kaempferol (− 9.60 kcal/mol) from P. amarus have exhibited better binding energies compared to remdesivir (− 9.50 kcal/mol). Apart from this, the binding affinities of furosin (− 9.20 kcal/mol) and geraniin (− 9.20 kcal/mol) are reasonably well with that of the positive control, remdesivir. Quercetin-3-O-glucoside has the maximum dock score of − 10.30 kcal/mol and shown its binding to HIS74, ARG83, TYR155, ASN157 and HIS176 amino acid residues of viral protein at the binding pocket of PLpro with eight hydrogen bonds (Table 2, Fig. 3).
The binding affinity of compounds from A. paniculata with PLpro ranged from -6.80 kcal/mol (for andrograpanin) to − 9.00 kcal/mol (for 5-Hydroxy-7, 8-dimethoxyflavone). Even though binding affinities of all the 15 compounds falls between -6.00 kcal/mol and less than the dock score of positive control remdesivir (− 9.5 kcal/mol), they also might have inhibitory effects on the activity of PLpro. The results suggest that, among the promising phytochemicals from P. amarus and A. paniculata docked with PLpro, flavonoids (quercetin-3-O-glucoside and astragalin) and phenylpropanoid (hinokinin) have exhibited better binding affinity. The SARS-CoV-2 PLpro is an essential for virus replication (Baez-Santos et al. 2014) and any compound having good binding affinity with inhibitory effects will be of immense potential in containing this virus. The flavonoids and phenylpropanoids showing good affinity binding abilities might serve as lead molecules for the discovery of novel drugs to SARS-CoV-2.
Interaction between RNA-dependent RNA polymerase (RdRp) (6NUS) and phytochemicals
The RdRp plays a major role in the replication and transcription of SARS-CoV-2 genome (Aftab et al. 2020). The RdRp used in the current study is the solved structure of SARS-CoV having 97.08 percent sequence identity with the RdRp of SARS-CoV-2 (Kirchdoeffer and Ward 2020).
Among 21 compounds from P. amarus screened against RdRp, epibubbialine has shown lowest dock score of − 6.30 kcal/mol (below − 6.0 kcal/mol is considered as having good binding affinity) and corilagin has shown highest dock score of − 9.30 kcal/mol (Table 1). Seven compounds: corilagin (− 9.30 kcal/mol), furosin (− 9.10 kcal/mol), geraniin (− 9.10 kcal/mol), quercetin (− 8.60 kcal/mol), hinokinin (− 8.20 kcal/mol), quercetin (− 8.10 kcal/mol) and quercetin-3-O-glucoside (− 7.70 kcal/mol) from P. amarus exhibited better dock score compared to the RdRp antagonist remdesivir (− 7.60 kcal/mol). Corilagin with high binding affinity of − 9.30 kcal/mol has interacted with active site residues, TYR129, HIS133, LEU207, ASN705 and GLN724 of RdRp of virus through five hydrogen bonds (Table 2, Fig. 3).
Five compounds, neoandrographolide (-8.50 kcal/mol), isoandrographolide (− 8.30 kcal/mol), 14-deoxy-14,15-didehydroandrographolide (− 7.90 kcal/mol), 14-Deoxyandrographolide (− 7.80 kcal/mol) and 5-hydroxy-7, 8, 2′,5′-tetramethoxy flavone (− 7.80 kcal/mol) from A. paniculata, have shown better binding energy affinity compared to remdesivir (− 7.60 kcal/mol) (Table 1). Among these, neoandrographolide exhibited higher dock score of − 8.50 kcal/mol and interacted with ALA125, ASP126, ARG132 and HIS133 amino acid residues of RdRp through four hydrogen bonds (Table 2, Fig. 3).
In general, the flavonoids (astragalin, kaempferol, quercetin, quercetin-3-O-glucoside and quercetin), a phenylpropanoid (hinkonin) and tannins (corilagin, furosin and geraniin) from P. amarus and a bioactive compound (isoandrographolide), diterpenes (14-deoxy-14,15-didehydroandrographolide, 14-Deoxyandrographolide, 19-O-acetyl-14-deoxy-11,12 didehydroandrographolide, neoandrographolide) and a flavonoid (5-hydroxy-7,8,2′,5′-tetramethoxy flavone) from A. paniculata have shown good binding affinities with all the four SARS-CoV-2 target proteins. The natural compounds obtained from the medicinal plants have been used in case of two previous coronavirus outbreaks of MERS-CoV and SARS-CoV (Li et al. 2005; Lin et al. 2005). Similarly, strong interaction of tannins (polyphenols) from green tea, epigallocatechin gallate, epicatechingallate and gallocatechin-3-gallate with viral receptor (3CLpro) of SARS-Cov-2 was shown (Ghosh et al. 2020). The flavonoids and tannins interaction with proteins of SARS-CoV-2 in vitro may provide more insights into exploiting these compounds as potential SARS-CoV-2 inhibiting compounds and lead to novel drug development. Further, among all the phytochemicals, the geraniin has shown consistent interaction and very good binding affinity with all the viral protein receptors. The geraniin, extracted from P. amarus, has shown to inhibit HIV-1 replication in HeLa CD4 + cells with 50% effective concentration (EC50) values ranging from 0.9 to 7.6 μg/ml (Notka et al. 2004). This suggests that geraniin with multiple target binding abilities in docking analysis with high dock score than remdesivir can be a potential candidate to harness as anti SARS-CoV-2 drug. However, this needs in vitro evaluation to authenticate the prediction studies.
ADME and other pharmacokinetic properties
The pharmacokinetic evaluation and ADME (absorption, distribution, metabolism and excretion) properties predictions of pharmacological compounds in advance are important methods to save huge monetary expenses in drug discovery (Zhu et al. 2011). Apart from that, it mainly helps in reducing the potential risk during clinical development as it provides very crucial information to determine whether a compound is suitable to proceed to clinical stage. SWISS-ADME (http://www.swissadme.ch) and admetSAR (http://lmmd.ecust.edu.cn/admetsar2) web servers were used to obtain the ADME and other pharmacokinetic properties of the selected compounds. Among the 35 compounds from P. amarus and A. paniculata evaluated against SARS-CoV-2 by in silico docking analysis, 15 compounds (9 compounds from P. amarus and 6 compounds from A. paniculata) were selected for ADME and other pharmacokinetic properties predictions. All these 15 compounds selected showed better binding ability than the remdesivir. The ADME properties of the selected 15 compounds have been represented in supplementary Fig. 2.
The ADME and pharmacokinetic properties of selected phytochemicals are given in Table 3. Lipinski rule of five (Lipinski 2004) is a thumb rule to evaluate the drug likeliness or to determine the pharmacological activities that would make it orally active drug in humans. According to the rule, the compound should abide following criteria, i.e., (1) molecular mass less than 500 Daltons, (2) high lipophilicity (expressed as logP less than 5), (3) less than five hydrogen bond donors and (4) less than ten hydrogen bond acceptors (Petit et al. 2012). Of 15 compounds along with remdesivir screened, eight compounds (kaempferol, quercetin, hinokinin, isoandrographolide, 14-deoxy-14, 15-didehydroandrographolide, 14-Deoxyandrographolide, 19-O-Acetyl-14-deoxy-11, 12-didehydroandrographolide and neoandrographolide) did not violate any of the above rule. However, four compounds, viz. astragalin (HBA > 10, HBD > 5), quercetin-3-O-glucoside (HBA > 10, HBD > 5), quercitrin (HBA > 10, HBD > 5) and 5-hydroxy-7, 8, 2′, 5′-tetramethoxy flavone (MW > 500, HBA > 10) along with positive control remdesivir, have violated two rules. Three compounds belong to tannins, i.e., corilagin (MW > 500, HBA > 10, HBD > 5), furosin (MW > 500, HBA > 10, HBD > 5) and geraniin (MW > 500, HBA > 10, HBD > 5), have violated three rules (Table 3). The violation of two or more criteria predicts a molecule to be a non-orally available drug (Lipinski 2004). However, this rule applies only for oral drugs, and the compounds violating the Lipinski’s rule of five can be administered through other means after determining its side effects (Benet et al. 2016).
Table 3.
Absorption, distribution, metabolism and excretion (ADME) and other pharmacokinetic properties of selected phytochemicals from Phyllanthus amarus and Andrographis paniculata
| S. L. No. | Group | Compound | HIA | Carcinogens | logS value | MW | HBD | HBA | logP value | Rule of 5 violations |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Positive control | Remdesivir | 0.9135 (low) | NC | − 3.474 | 602.58 | 4 | 12 | 3.4 | 2 |
| 2 | Flavonoids | Astragalin | 0.6347 (low) | NC | − 2.449 | 448.38 | 7 | 11 | 1.29 | 2 |
| 3 | Kaempferol | 0.9881 (high) | NC | − 3.142 | 286.24 | 4 | 6 | 1.7 | 0 | |
| 4 | Quercetin | 0.9883 (high) | NC | − 2.999 | 302.24 | 5 | 7 | 1.63 | 0 | |
| 5 | quercetin-3-O-glucoside | 0.6468 (low) | NC | − 2.449 | 464.38 | 8 | 12 | 0.94 | 2 | |
| 6 | Quercitrin | 0.7322 (low) | NC | − 3.497 | 448.38 | 7 | 11 | 1.6 | 2 | |
| 7 | Phenylpropanoid | Hinokinin | 0.9784 (high) | NC | − 3.113 | 354.35 | 0 | 6 | 3.08 | 0 |
| 8 | Tannins | corilagin | 0.8598 (low) | NC | − 2.727 | 634.45 | 11 | 18 | 1.48 | 3 |
| 9 | Furosin | 0.8598 (low) | NC | − 2.75 | 634.45 | 11 | 18 | 1.48 | 3 | |
| 10 | geraniin | 0.8592 (low) | NC | − 3.172 | 954.66 | 14 | 27 | 0.67 | 3 | |
| 11 | Bioactive compound | Isoandrographolide | 0.9822 (high) | NC | − 2.853 | 350.45 | 3 | 5 | 2.45 | 0 |
| 12 | Diterpenes | 14-deoxy-14,15-didehydroandrographolide | 0.9773 (high) | NC | − 2.927 | 332.43 | 2 | 4 | 2.96 | 0 |
| 13 | 14-Deoxyandrographolide | 0.9899 (high) | NC | − 2.816 | 334.45 | 2 | 4 | 2.79 | 0 | |
| 14 | 19-O-Acetyl-14-deoxy-11,12-didehydroandrographolide | 0.9764 (high) | NC | − 4.139 | 374.47 | 1 | 5 | 3.48 | 0 | |
| 15 | Neoandrographolide | 0.8124 (low) | NC | − 3.313 | 480.59 | 4 | 8 | 3.37 | 0 | |
| 16 | Flavonoids | 5-hydroxy-7,8,2′,5′-tetramethoxy flavone | 0.4104 (low) | NC | − 2.678 | 520.48 | 4 | 12 | 3.22 | 2 |
HIA human intestinal absorption (probability), logS solubility, MW molecular weight (Dalton), HBD hydrogen bond donor, HBA hydrogen bond acceptor, logP liphophilicity, Rule of 5 violations Lipinski rule of 5 and NC-Non carcinogen
The logS value is directly related to the solubility of the drug in water which ideally ranged between − 6.5 and 0.5 (Nisha et al. 2016). All the screened compounds lie within this range. The solubility of the molecules in water significantly facilitates drug developmental activities (Diana et al. 2017). The selected phytochemicals have passed the carcinogenic test and all exhibit non-carcinogenic nature (Table 3). Among 15 phytochemicals, seven phytochemicals (kaempferol, quercetin, hinokinin, isoandrographolide, 14-deoxy-14,15-didehydroandrographolide, 14-Deoxyandrographolide and 19-O-acetyl-14-deoxy-11, 12-didehydroandrographolide) have shown high gastrointestinal or human intestinal absorption. Remaining eight phytochemicals (astragalin, quercetin-3-O-glucoside, quercetin, corilagin, furosin, geraniin, neoandropholidae and 5-hydroxy-7, 8, 2′, 5′-tetramethoxy flavone) have exhibited low gastrointestinal or human intestinal absorption. The positive control remdesivir also has low human intestinal absorption value. As all the compounds have exhibited non-carcinogenic nature and good pharmacokinetic properties these compounds individually or in combination may be effective against the SARS-CoV-2. Further, inflammation of lungs resulting in breathing problem is a major symptom in the critical patients suffering from Covid-19 (Li and Ma 2020). In addition to all the prediction studies, the plant extracts of P. amarus have shown anti-inflammatory properties in rat Kupffer cells (Kiemer et al. 2003). The pharmacokinetic predictions suggest that compounds from the two plants species studied in the current study are found to be biologically safe and can be utilized as therapeutics against SARS-CoV-19. However, more studies need to be conducted to decipher the exact mechanism involved in the interaction between these phytochemicals and the viral receptor proteins.
Conclusion
To summarize, all these current studies were done by in silico predictions of individual phytochemical originated from P. amarus and A. paniculata. The information generated here may provide some insights into exploring for potential drugs against SARS-CoV-2 from plants, which can result in discovery of novel drugs. Further, the Ayurveda medications using herbal extracts cannot be undermined in addressing human ailments, because these extracts contain several compounds, which might have synergistic effect with action on multi-target sites of pathogen. However, the biggest problem in Ayurveda medicine is fixing the dose against the target pathogens. Optimization of this issue supported by scientific evidences for getting required clearances which will have far fetching impact in the harnessing Indian ancient knowledge of medication “Ayurveda” in treating the human ailments including Covid-19 caused by SARS-CoV-2, which threatened the mankind with global pandemic outbreak.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
SH performed the bioinformatics work, SH, MN, HDV, MM, KSS and CNL involved in experimental design, CNL conceptualized work and provided the overall direction, all authors are involved literature mining and manuscript preparation. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Contributor Information
Shridhar Hiremath, Email: shridharhi80@gmail.com.
H. D. Vinay Kumar, Email: hargi.vinay@gmail.com.
M. Nandan, Email: nandaaa94@gmail.com
M. Mantesh, Email: manteshmuttappagol@gmail.com
K. S. Shankarappa, Email: ksshankarappa@gmail.com
V. Venkataravanappa, Email: venkatrajani@gmail.com
C. R. Jahir Basha, Email: jahir_basha@rediffmail.com.
C. N. Lakshminarayana Reddy, Email: cnlreddy@gmail.com.
References
- Adem S, Eyupoglu V, Sarfraz I, Rasul A, Ali M. Identification of Potent COVID-19 Main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against Corona. Preprints. 2020 doi: 10.20944/preprints202003.0333.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aftab SO, Ghouri MZ, Masood MU, Haider Z, Khan Z, Ahmad A, Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020;18(1):1–15. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos YM, et al. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57(6):2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum J, Mir NA, Dev K, Buyamayum B, Wani MY, Raza M (2020) Challenges and prospects of COVID‐19 vaccine development based on the progress made in SARS and MERS vaccine development. Transbound Emerg Dis. 10.1111/tbed.13804 [DOI] [PMC free article] [PubMed]
- Benet LZ, Hosey CM, Ursu O, Oprea TI. BDDCS, the rule of 5 and drugability. Adv Drug Delivery Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia DS (2020) Discovery studio visualizer, Release 2020, San Diego: Dassault Systemes
- Boopathi S, Poma AB, Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto JB, Santos AR, Filho VC, Yunes RA. A review of the plants of the genus Phyllanthus: their chemistry, pharmacology, and therapeutic potential. Med Res Rev. 1998;18(4):225–258. doi: 10.1002/(sici)1098-1128(199807)18:4<225::aid-med2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with a typical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YS, Woo ER. Korean medicinal plants inhibiting to Human Immunodeficiency Virus type 1 (HIV-1) fusion. Phytother Res. 2003;17(4):426–429. doi: 10.1002/ptr.1155. [DOI] [PubMed] [Google Scholar]
- Chao WW, Lin BF. Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian) Chin Med. 2010;5(1):17. doi: 10.1186/1749-8546-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Qiu H, Gong J, Liu Q, Xiao H, Chen XW, Sun BL, Yang RG. Epigallocatechin-3-gallate inhibits the replication cycle of hepatitis C virus. Arch Virol. 2012;157(7):1301–1312. doi: 10.1007/s00705-012-1304-0. [DOI] [PubMed] [Google Scholar]
- Chen D, Oezguen N, Urvil P, Ferguson C, Dann SM, Savidge TC. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci Adv. 2016;2(3):1501240. doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and re-emerging infection. Clin Microbiol Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir, Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2009) The PyMOL Molecular Graphics System http://www.pymol.org
- Diana M, Raij T, Melis M, Nummenmaa A, Leggio L, Bonci A. Rehabilitating the addicted brain with transcranial magnetic stimulation. Nat Rev Neurosci. 2017;18(11):685. doi: 10.1038/nrn.2017.113. [DOI] [PubMed] [Google Scholar]
- Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;2020(248):117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani M, Teimoori A, Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol. 2020;15(5):317–323. [Google Scholar]
- Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In Coronaviruses, Humana Press, New York, pp 1-23 [DOI] [PMC free article] [PubMed]
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gescher K, Hensel A, Hafezi W, Derksen A, Kühn J. Oligomeric proanthocyanidins from Rumex acetosa L. inhibit the attachment of herpes simplex virus type-1. Antivir Res. 2011;89(1):9–18. doi: 10.1016/j.antiviral.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomathi M, Padmapriya S, Balachandar V. Drug studies on Rett syndrome: from bench to bedside. J Autism Dev Disord. 2020;50:2740–2764. doi: 10.1007/s10803-020-04381-y. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Vaghela JS. Recent advances in pharmacological and phytochemistry studies on Phyllanthus amarus. J Pharm Biosci. 2019;7(1):1–8. [Google Scholar]
- Gupta S, Mishra K, Ganju L. Broad-spectrum antiviral properties of andrographolide. Arch Virol. 2017;162(3):611–623. doi: 10.1007/s00705-016-3166-3. [DOI] [PubMed] [Google Scholar]
- Gur M, Taka E, Yilmaz SZ, Kilinc C, Aktas U, Golcuk M. Conformational transition of SARS-CoV-2 spike glycoprotein between its closed and open states. The J Chem Phys. 2020;153(7):075–101. doi: 10.1063/5.0011141. [DOI] [PubMed] [Google Scholar]
- Hall DC, Jr, Ji HF. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Di. 2020;35:101–646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminf. 2012;4(1):1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;9:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhibition Med Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Raj SJ. An overview: pharmacognostic properties of Phyllanthus amarus Linn. Int J Pharmacol. 2011;7(1):40–45. [Google Scholar]
- Karakus U, Pohl MO, Stertz S. Breaking the convention: sialoglycan variants, coreceptors, and alternative receptors for influenza a virus entry. J Virol. 2020;94(4):1–9. doi: 10.1128/JVI.01357-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Bioph Res Co. 2004;323(1):264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer AK, Hartung T, Huber C, Vollmar AM. Phyllanthus amarus has anti-inflammatory potential by inhibition of iNOS, COX-2, and cytokines via the NF-κB pathway. J Hepatol. 2003;38(3):289–297. doi: 10.1016/s0168-8278(02)00417-8. [DOI] [PubMed] [Google Scholar]
- Kim Y, Narayanan S, Chang KO. Inhibition of influenza virus replication by plant-derived isoquercetin. Antivir Res. 2010;88(2):227–235. doi: 10.1016/j.antiviral.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11(13):580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Kumari K, Jayaraj A, Kumar V, Kumar RV, Dass SK, Chandra R, Singh P. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1752310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? J Crit Care. 2020;24(198):1–5. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, Zhang X, Hua SN, Yu J, Xiao PG, Li RS. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Tsai FJ, Tsai CH, Lai CC, Wan L, Ho TY, Hsieh CC, Chao PDL. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir, Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machhi J, Herskovitz J, Senan AM. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J Neuroimmune Pharmacol. 2020 doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal B. The global emergence of severe acute respiratory syndrome coronavirus 2 in human. Virus dis. 2020;31:67–70. doi: 10.1007/s13337-020-00613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri S, Choudhary G, Sarzala PM, Ratner L, Hudak KA. Suppression of human T-cell leukemia virus I gene expression by pokeweed antiviral protein. J Biol Chem. 2009;284(45):31453–31462. doi: 10.1074/jbc.M109.046235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan NA, Pandian CJ, Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1777901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjan A, Tewari SK, Lehri A. Biological activities of kalmegh (Andrographis paniculata Nees) and its active principles: a review. Indian J Nat Prod Resour. 2010;1(2):125–135. [Google Scholar]
- Nisar MF, He J, Ahmed A, Yang Y, Li M, Wan C. Chemical components and biological activities of the genus Phyllanthus: a review of the recent literature. Molecules. 2018;23(10):2567. doi: 10.3390/molecules23102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisha CM, Kumar A, Nair P, Gupta N, Silakari C, Tripathi T, Kumar A. Molecular docking and in silico ADMET study reveals acylguanidine 7a as a potential inhibitor of β-secretase. Adv Bioinform. 2016 doi: 10.1155/2016/9258578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notka F, Meier G, Wagner R. Concerted inhibitory activities of Phyllanthus amarus on HIV replication in vitro and ex vivo. Antivir Res. 2004;64(2):93–102. doi: 10.1016/j.antiviral.2004.06.010. [DOI] [PubMed] [Google Scholar]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Chem inf. 2011;3(1):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P, Rane JS, Chatterjee A, Kumar A, Khan R, Prakash A, Ray S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Tripathi P, Sharma V, Chauhan NS, Dixit VK. Phyllanthus amarus: ethnomedicinal uses, phytochemistry and pharmacology: a review. J Ethnopharmacol. 2011;138(2):286–313. doi: 10.1016/j.jep.2011.09.040. [DOI] [PubMed] [Google Scholar]
- Pereira RG, Garcia VL, Rodrigues MVN, Martínez J. Extraction of lignans from Phyllanthus amarus Schum. & Thonn using pressurized liquids and low pressure methods. Sep Purif Technol. 2016;158:204–211. [Google Scholar]
- Petit J, Meurice N, Kaiser C, Maggiora G. Softening the rule of five-where to draw the line. Bioorg Med Chem. 2012;20(18):5343–5351. doi: 10.1016/j.bmc.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pholphana N, Rangkadilok N, Saehun J, Ritruechai S, Satayavivad J. Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm. F.) Nees (Chuanxinlian) Chin. Med. 2013;8(1):1–12. doi: 10.1186/1749-8546-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussia A, Thepchatri P, Snyder JP, Plemper RK. Systematic approaches towards the development of host-directed antiviral therapeutics. Int J Mol Sci. 2011;12(6):4027–4052. doi: 10.3390/ijms12064027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar MT, Alqahtani SM, Alamri MA, Chen LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, Montgomery SA, Hogg A, Babusis D, Clarke MO, Spahn JE. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shityakov S, Sohajda T, Puskas I, Roewer N, Forster C, Broscheit JA. Ionization states, cellular toxicity and molecular modeling studies of midazolam complexed with trimethyl-β-cyclodextrin. Molecules. 2014;19(10):16861–16876. doi: 10.3390/molecules191016861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MCS, Oyong GG, Shen CC, Ragasa CY. Chemical constituents of Andrographis paniculata (Burm. F.) Nees. Int J Pharmacogn Phytochem Res. 2016;8(8):1398–1402. [Google Scholar]
- Thayil SM, Thyagarajan SP. Pa-9: a flavonoid extracted from Plectranthus amboinicus inhibits HIV-1 protease. Int J Pharmacogn Phytochem Res. 2016;8(6):1020–1024. [Google Scholar]
- Tortorici MA, Veesler D (2019) Structural insights into coronavirus entry. In Adv Virus Res 105: 93–116 [DOI] [PMC free article] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Burton DR. Passive immunotherapy of viral infections, ‘super-antibodies’ enter the fray. Nat Rev Immunol. 2018;18(5):297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Y, Ye D, Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Chen L, Lan R, Shen R, Li P (2020) Computational screening of antagonist against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents, 106012 [DOI] [PMC free article] [PubMed]
- Zakaryan H, Arabyan E, Oo A, Zandi K. Flavonoids: promising natural compounds against viral infections. Arch Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang J, Yu H, Li Y, Hou T. Recent developments of in silico predictions of oral bioavailability. Comb Chem High Throughput Screen. 2011;14(5):362–374. doi: 10.2174/138620711795508368. [DOI] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.