Abstract
White-cheeked macaque is a newly described species in genus Macaca. Here, the complete mitochondrial genome was firstly determined, which was deposited in Genbank with accession number KU564271. The length of the mitogenome is 16,494 base pairs (bp). It includes 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and a control region. We used 12 mitochondrion genes of 15 species to constructed phylogenetic tree with three methods. The study will provide useful data for further studies on phylogenetic relationships as well as population structure, biodiversity conservation and conservation genetics.
Key words: Genome structure, Macaca leucogenys, mitochondrial genome, phylogenetic tree
The white-cheeked macaque (Macaca leucogenys) was described as a new macaque species in Medog, southeastern Tibet, China (Li et al. 2015). Little studies have been done on molecule biology and genetics of M. leucogenys. Here, we sequenced the complete mitochondrion genome of M. leucogenys and 12 genes of 14 species were used to construct the phylogenetic tree, which will not only facilitate the phylogenetic estimation but also provide valuable data for studies of population structure and conservation genetics.
The specimen was collected from Medog, Tibet, China, which is stored in Dali University, Yunnan province and preserved at low temperature (−20 °C) with the accession number is XKL. The total genomic DNA was extracted with the method of standard phenol/chloroform (Sambrook & Russell 2001). The complete mitogenome of M. leucogenys is 16,494 bp, which was deposited in the Genbank with the accession number KU564271. It contains 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and a control region. The base composition of complete mitochondrial genome is 31.44% A, 25.26% T, 30.08% C and 13.21% G. The content of A + T is 56.71%, which similar with other vertebrate mitochondrial genome (Zhang et al. 2003).
The PCGs regions of the M. leucogenys mitogenomes were consistent with those of other cercopithecidae mitogenomes (Boore 1999; Xing et al. 2007). Among the 13 PCGs, all the PCGs were located on H strand except ND6 genes, which was encoded on L strand. The majority of the genes were initiated with ATG. Three genes were initiated with ATN. However, ND1 used infrequently GTG as the start codon. Five PCGs used the standard codon TAA as stop codon, while ND2, COXI and CYTB used TAN as their incomplete stop codon. Single T was used as incomplete stop codon in ND1, COXIII, ND3, ND4 and ND6.
Macaca leucogenys contained the typical domains of 22 tRNA genes, interspersing in the genome and ranging in size from 59 bp to 75 bp, in line with what have been found in most cercopithecidae (Juhling et al. 2012). The repeat region is longer than the M. thibetana. As the same as other cercopithecidae species, eight tRNA encoded on the L strand, while the other 14 encoded on H strand. Besides, we found that tRNA-Glu gene in M. leucogenys is longer than the other species (i.e. M. thibetana, M. mulatta, etc.). A control gene (D-loop) is 1065 bp with the typical gene order in vertebrates.
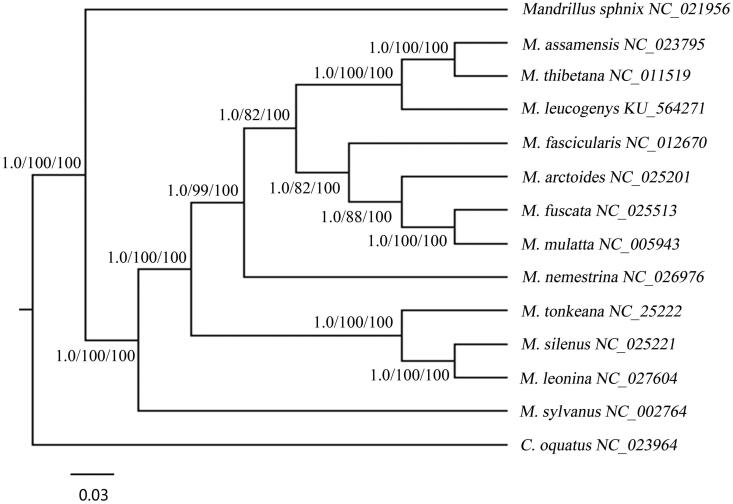
Macaca leucogenys is more similar to the silenus–sylvanus group, rather than the potential sympatric macaque species based on the structure of male external genitalia and morphology (i.e. M. assamensis and M. mulatta) (Li et al. 2009; Jiang et al. 2016). Phylogenetic analysis based on 12 common protein sequences except ND6 in 15 species. Three methods (BI/MP/NJ) were used to constructed the phylogenetic tree (Figure 1), which confirmed that M. leucogenys as a member of genus Macaca. Macaca leucogenys is most closely to the species M. assamensis and M. thibetana analyzed from the tree. It provides useful data for further study on evolution of M. leucogenys.
Figure 1.
Three methods were used to construct the phylogenetic tree, Bayes tree, maximum parsimony method and neighbour-joining method, which constructed by MrBayes (MrBayes Inc., New York, NY), PAUP4.0 (Sinauer Associates, Sunderland, MA) and MEGA5.2 (MEGA Inc., Englewood, NJ) individually. Macaca leucogenys is closely to Macaca assamensis and Macaca thibetana. Cerecocebus oquatus species was set as the outgroup.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Funding information
This research was supported by National Natural Science Foundation of China (Nos. 31270431 and 31530068).
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Li P, Yu J, Zhao G, Yi Y, Yue B, Li J. 2016. The complete mitochondrial genome of Assamese Macaques (Macaca assamensis). Mitochondrial DNA. 27:226–227. [DOI] [PubMed] [Google Scholar]
- Juhling F, Putz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. 2012. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40:2833–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhao C, Fan PF. 2015. White-cheeked macaque (Macaca leucogenys): a new macaque species from Medog, southeastern Tibet. Am J Primatol. 77:753–766. [DOI] [PubMed] [Google Scholar]
- Li D, Fan L, Zeng B, Yin H, Zou F, Wang H, Meng Y, King E, Yue B. 2009. The complete mitochondrial genome of Macaca thibetana and a novel nuclear mitochondrial pseudogene. Gene. 429:31–36. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Press. [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. 2007. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 134:2–19. [DOI] [PubMed] [Google Scholar]
- Zhang P, Chen YQ, Liu YF, Zhou H, Qu LH. 2003. The complete mitochondrial genome of the Chinese giant salamander, Andrias davidianus (Amphibia: Caudata). Gene. 311:93–98. [DOI] [PubMed] [Google Scholar]