Abstract
Eclipta prostrata is an herbal medicinal plant belonging to the Asteraceae family. In this study, complete chloroplast genome sequence of the E. prostrata was characterized by de novo assembly using whole genome sequence data. The genome of E. prostrata was 151,757 bp in length, which was composed of large single copy region of 83,285 bp, small single copy region of 18,346 bp and a pair of inverted repeat regions of 25,063 bp. The genome harboured 80 protein coding sequences, 30 tRNA genes and 4 rRNA genes. We confirmed close taxonomic relationship between E. prostrata and Helianthus annuus through phylogenetic analysis with chloroplast protein-coding genes.
Keywords: Chloroplast, Eclipta prostrata, genome sequence
Eclipta prostrata is an annual herbal plant species belonging to the Asteraceae family, which is mainly distributed in tropical and subtropical regions in the worlds (Panero et al. 1999). E. prostrata has been used as a folk medicine for treatment of inflammatory diseases such as hepatitis (Han et al. 1998). In addition, pharmacological studies reported various biological activities of E. prostrata, including anti-inflammatory and anticancer-cytotoxic activity (Arunachalam et al. 2009; Khanna & Kannabiran 2009; Liu et al. 2012). However, there is rare genetic and genomic study for authentication and breeding of these plants. In this study, we characterized chloroplast genome sequence of E. prostrata to provide useful genome information for this plant species.
To determine chloroplast genome sequence, we extracted total genomic DNA from leaves of E. prostrata which were provided by HanTaek Botaniccal Garden (Yongin, Korea, www.hantaek.co.kr). The extracted DNA was used to construct Ilumina paired-end (PE) genomic library and sequenced using an Illumina MiSeq platform (Illumina, San Diego, CA) in Lab Genomics Co. (Seongnam, Korea; http://www.labgenomics.co.kr) . High quality PE reads of ∼1.4 Gb were assembled by CLC genome assembler (ver. 4.06 beta, CLC Inc, Aarhus, Denmark) according to the previous study (Kim et al. 2015a, b), and then representative chloroplast contigs were retrieved, ordered and combined into a single draft sequence by comparing chloroplast genome sequence of Centaurea diffusa (KJ690264). The draft sequence was manually corrected by PE read mapping. The genes in the chloroplast genome were annotated using the DOGMA program (Wyman et al. 2004) and manual curation based on BLAST searches.
The complete chloroplast genome of E. prostrata (KU361242) was 151,757 bp in length, which was composed of large single copy region of 83,285 bp, small single copy region of 18,346 bp and a pair of inverted repeat regions of 25,063 bp, as typical chloroplast structure. A total of 114 genes were predicted in the genome, including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes.
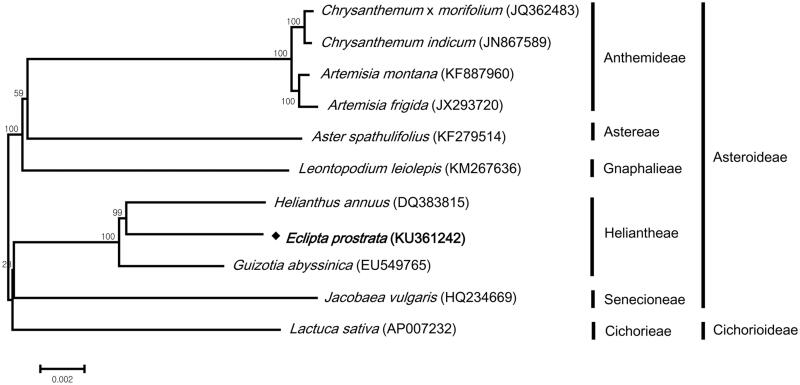
Phylogenetic analysis was performed using common 68 protein-coding genes in 11 chloroplast genomes by maximum likelihood method in the MEGA 6.0 (Tamura et al. 2013) for E. prostrata and related 10 species belonging to the Asteraceae family. The phylogenetic tree showed that E. prostrata was grouped with other two species in Heliantheae tribe, in which E. prostrata was placed more closely to Helianthus annuus (sunflower) (Figure 1).
Figure 1.
Phylogenetic tree showing relationship between Eclipta prostrata and 10 species belonging to the Asteraceae family. Phylogenetic tree was constructed based on 68 protein-coding genes of chloroplast genomes using maximum likelihood method with 1000 bootstrap replicates. Numbers in each the node indicated the bootstrap support values.
Acknowledgments
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
Funding information
This work was supported by “15172MFDS246” from Ministry of Food and Drug Safety in 2015, Republic of Korea.
References
- Arunachalam G, Subramanian N, Pazhani GP, Ravichandran V. 2009. Antiinflammatory activity of methanolic extract of Eclipta prostrata L. (Astearaceae). Afr J Pharm Pharmacol. 3:97–100. [Google Scholar]
- Han Y, Xia C, Cheng X, Xiang R, Liu H, Yan Q, Xu D. 1998. [Preliminary studies on chemical constituents and pharmacological action of Eclipta prostrata L]. Mater Med. 23:680–682. [PubMed] [Google Scholar]
- Khanna VG, Kannabiran K. 2009. Anticancer-cytotoxic activity of saponins isolated from the leaves of Gymnema sylvestre and Eclipta prostrata on Hela cells. Int J Green Pharm. 3:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLos One. 10:e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QM, Zhao HY, Zhong XK, Jiang JG. 2012. Eclipta prostrata L. phytochemicals: isolation, structure elucidation, and their antitumor activity. Food Chem Toxicol. 50:4016–4022. [DOI] [PubMed] [Google Scholar]
- Panero JL, Jansen RK, Clevinger JA. 1999. Phylogenetic relationships of subtribe Ecliptinae (Asteraceae: Heliantheae) based on chloroplast DNA restriction site data. Am J Bot. 86:413–427. [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 . Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]