Abstract
Magic Lily (Lycoris squamigera), belonging to the Amaryllidaceae family, is cultivated for ornamental and medicinal purposes. To characterize its genomic information, we obtained the complete chloroplast genome sequence of L. squamigera by assembling Illumina whole genome sequence data. The complete chloroplast genome is 158,482 bp in length which is composed of four unique regions, a large single copy region (LSC) of 86,454 bp, a small single copy region (SSC) of 18,500 bp, and a pair of inverted repeats (IR) of 26,764 bp. The genome annotation predicted 159 genes including 105 protein-coding genes, 46 tRNA genes, and 8 rRNA genes. Phylogenetic tree analysis revealed that L. squamigera clustered with Allium species belonging to the Amaryllidaceae family.
Keywords: Chloroplast, genome sequence, Lycoris squamigera
Magic Lily (Lycoris squamigera) belongs to the Amaryllidaceae family, which contains more than 750 species (Friesen et al. 2006). It has been cultivated not only for its ornamental value but also for its pharmacologically active alkaloids such as galanthine and squamigine (Martin 1987, Jin et al. 2011). Although there are many studies focusing on their ingredients, there are few studies conducted on their genome including three complete chloroplast genomes in the Amaryllidaceae family (NC_035971.1, NC_031829.1; Filyushin et al. 2016 and KX683282; Kim et al. 2016). Due to sharing of similar phenotypes between Lycoris genus, classification among this genus is difficult (Yoo et al. 2011). Since chloroplast genomes are widely used for understanding genetic diversity and evolution of plants (Nguyen et al. 2018; Joh et al. 2017), understanding the chloroplast genome has great significance. In this study, we report the complete chloroplast genome sequence of L. squamigera by assembling whole-genome Illumina sequence data to understand its genome and provide concrete information on the phylogenetic relationship of this family.
The leaves of L. squamigera were collected from Hantaek Botanical Garden (Yongin, Republic of Korea, 37°05'41.6″N 127°24'23.4″E) for preparation of whole genome DNA. Sequencing was conducted by the Illumina Miseq Sequencing platform (Illumina, CA), and 1.3 Gb of sequence data was generated. Quality trimming and assembly of the reads were conducted by the de novo assembly of low coverage whole genome sequence (dnaLCW method) (Kim et al. 2015a,b) using a CLC genome assembler version 4.21 (CLC Inc., Aarhus, Denmark). Assembled contigs were joined into a single draft sequence by guidance of chloroplast genome of Allium cepa (KF28079) as a reference. Confirmation and correction of the draft sequence were manually done with mapping of raw reads and BLAST searches. The plastid genome was annotated with GeSeq (Tillich et al. 2017) and manually curated.
The total chloroplast genome size of L. squamigera was 158,482 bp (GenBank accession no. MH118290) which is composed of four typical chloroplast regions: a large single copy region (LSC) of 18,500 bp, a small single copy region (SSC) of 26,764 bp, and a pair of inverted repeat region of 26,764 bp. The plastid genome contains 159 genes including 105 protein-coding genes, 46 tRNA genes, and 8 rRNA genes. All of the genes are single copy genes excluding 25 protein-coding genes duplicated in the IR regions.
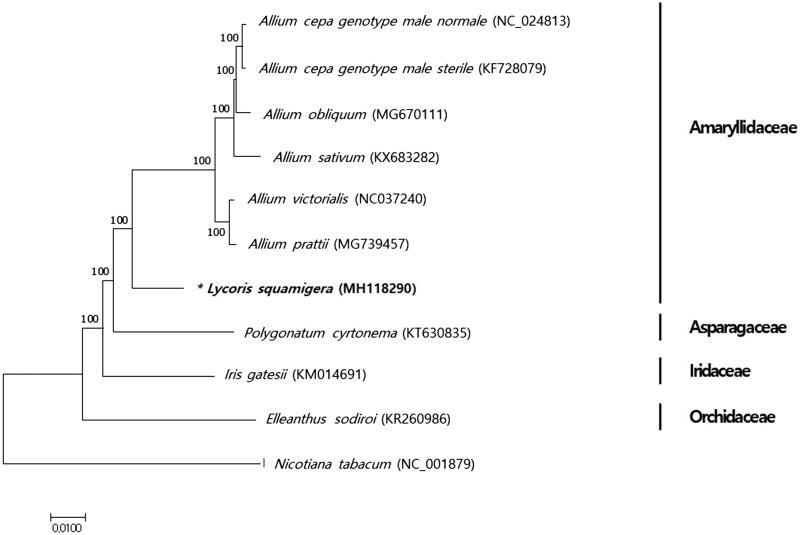
Phylogenetic analysis was performed with coding sequences of L. squamigera including eight chloroplast genomes previously reported in Asparagales using a neighbour-joining analysis of MEGA 7 (Kumar et al. 2016) with 1000 bootstrap replicates. All species were classified correctly into four groups: Amaryllidaceae, Orchidaceae, Asparagaceae, and Iridaceae (Figure 1). In this phylogenetic tree, L. squamigera was grouped together with six Allium species in the Amaryllidaceae family.
Figure 1.
Phylogenetic relationship of L. squamigera with other 10 species of Asparagales. The tree was generated using 82 protein-coding gene sequences and full chloroplast genome sequences based on the neighbor-joining analysis of MEGA7. Numbers next to the tree indicate the bootstrap value from 1000 replicates. The sequence of Nicotiana tabacum was set as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Filyushin MA, Beletsky AV, Mazur AM, Kochieva EZ. 2016. The complete plastid genome sequence of garlic Allium sativum L [Article]. Mitochondrial DNA Part B-Resour. 1:831–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen N, Fritsch RM, Blattner FR. 2006. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA its sequences. Aliso. 22:372–395. [Google Scholar]
- Jin Z. 2011. Amaryllidaceae and sceletium alkaloids. Nat Prod Rep. 28:1126–1142. [DOI] [PubMed] [Google Scholar]
- Joh HJ, Kim NH, Jayakodi M, Jang W, Park JY, Kim YC, In JG, Yang TJ. 2017. Authentication of golden-berry P. ginseng cultivar ‘gumpoong’from a landrace ‘hwangsook’ based on pooling method using chloroplast-derived markers. Plant Breed Biotechnol. 5:16–24. [Google Scholar]
- Kim B, Kim K, Yang TJ, Kim S. 2016. Completion of the mitochondrial genome sequence of onion (Allium cepa L.) containing the CMS-S male-sterile cytoplasm and identification of an independent event of the ccmF N gene split]. Curr Genet. 62:873–885. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SF. 1987. The Amaryllidaceae alkaloids In: Brossi A, editor. The alkaloids: chemistry and pharmacology. Cambridge (MA): Academic Press; Chapter 3, p. 251–376. [Google Scholar]
- Nguyen VB, Giang VNL, Waminal NE, Park HS, Kim NH, Jang W, Lee J, Yang TJ. 2018. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique SNP markers. J Ginseng Res.doi: 10.1016/j.jgr.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YK, Yuan T, Lee JS, Lee AK, Roh MS, Kurita S, Suh JK. 2011. Species relationships of Lycoris endemic to Korea evaluated by RAPD and SNPs of nrDNA-ITS regions. Hortic Environ Biotechnol. 52:145–151. [Google Scholar]