Abstract
In this paper, the complete mitochondrial DNA (mtDNA) sequence of Thryssa kammalensis was determined. The mitochondrial genome is 16,884 bp in length, including 13 protein-coding genes, 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and a non-coding control region as those found in other vertebrates, with the gene identical to that of typical vertebrates. The overall base composition of the heavy strand is 31.07% A, 24.82% T, 28.33% C, and 15.78% G, with an AT bias of 55.89%. Phylogeny of T. kammalensis suggested more close relationship with Lycothrissa crocodilus and Setipinna taty. The complete mitogenome of this species can provide a basic data for the studies on population history, molecular systematics, phylogeography, stock evaluation, and conservation genetics.
Keywords: Engraulidae, molecular systematics, mitochondrial genome, Thryssa kammalensis
Thryssa kammalensis (Madura anchovy) is a commercially important Clupeiformes fish species in China, which belongs to the family Engraulidae and is distributed in the western part of the Indian Ocean and the Pacific Ocean. Mitochondrial DNA is used as a genetic marker for species identification, genetic structures, systematic, and phylogeography in various organisms due its maternal inheritance, high mutation rate, and rapid evolution (Avise et al. 1987). In this study, the specimens of T. kammalensis were collected from Shanwei, Guangdong Provience, China. The specimen was stored in the Key Laboratory of Aquatic Genetic Resources and Utilization (AGRU) of the Ministry of Agriculture in Shanghai Ocean University, and named ‘TK01’ as sample code. The genomic DNA was extracted by modified PCI method followed by ethanol precipitation (Sambrook & Russel 2001). Primers were designed on the basis of mitogenome sequences of Setipinna taty (GenBank Accession No. KC439458), a closely related species of T. kammalensis (Lavoue et al. 2010). The complete mitochondrial DNA sequence of T. kammalensis was firstly determined (GenBank Accession No. KU761588) using PCR and sequencing. According to our data, the complete mitochondrial genome of T. kammalensis was 16,884 bp in length, including 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, 1 L-strand replication origin (OL), and one control region (D-loop). The overall nucleotide composition was 31.07% A, 24.82% T, 28.33% C, and 15.78% G, with an AT bias of 55.89% which is similar to other fishes (Cheng et al. 2010, 2011a; Jin et al. 2012). The order, direction, locations of the genes and gene coding strand in the mitogenome were nearly identical to those in other Engraulidae species (Zhang & Sun 2013; Li et al. 2014; Zhang & Xian 2014) and the usual vertebrate consensus (Xu et al. 2011; Cheng et al. 2011b; Shi et al. 2012). ND6 gene and eight tRNA genes were encoded on the light(L) strain and the remaining genes were encoded on the heavy (H) strain. For protein-coding genes, 12 were initiated with the orthodox ATG except for COI with GTG, and the three types of termination codons revealed were TAA (ND1, ND4L, ND5, COI, ATP8, ATP6, COIII, ND3 and ND6), T(COII, Cytb, and ND4), TAG(ND2). The incomplete stop codons were presumably completed by post-transcriptional polyadenylation (Ojala et al. 1981). All the tRNA genes of T. kammalensis fold into a typical cloverleaf structure, except tRNAser (AGY) which lacks a dihydrouridine arm identified by tRNA scan SE 1.21 (Lowe & Eddy 1997). The two ribosomal RNA genes, 12S rRNA (949bp) and 16S rRNA (1681bp), are located between tRNAPhe and tRNALeu (UUR) and are separated by tRNAVal gene. The control region is located between tRNApro and tRNAphe and is 1223 bp in length.
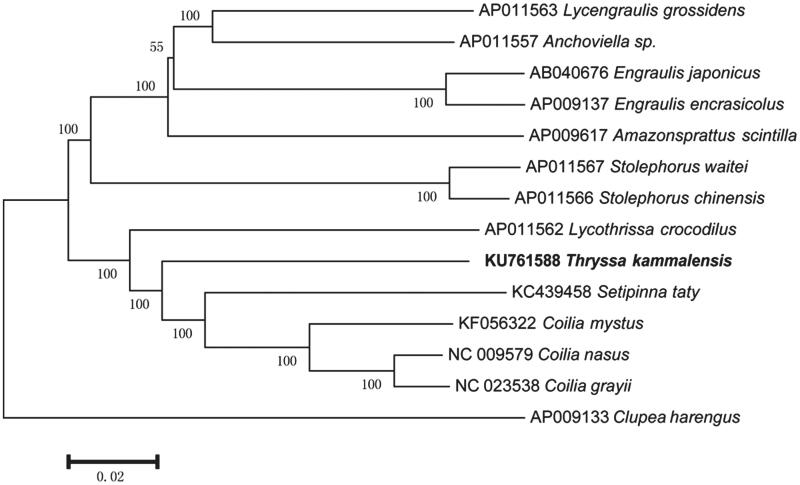
Phylogenetic analysis was done using MEGA 6.0 (Tamura et al. 2013) program with 13 complete mitogenomes under family Engraulidae and one sequence of Clupea harengus as the out-group. We found that the phylogeny of T. kammalensis suggested more close relationship with Lycothrissa crocodilus and S. taty, consistent with the traditional evolutionary position (Figure 1).
Figure 1.
Neighbour-joining (NJ) tree showing relationships among Thryssa kammalensis and related families.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Avise JC, Arnold J, Ball RM, Bermingham E, Lambt T, Neigel JE, Reeb CA, Saunders NC. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systemics. Ann Rev Ecol. 18:489–522. [Google Scholar]
- Cheng YZ, Wang RX, Xu TJ. 2011a. The mitochondrial genome of the spinyhead croaker Collichthys lucida: genome organization and phylogenetic consideration. Mar Genomics. 4:17–23. [DOI] [PubMed] [Google Scholar]
- Cheng YZ, Xu TJ, Jin XX, Wang RX. 2011b. Complete mitochondrial genome of the yellow drum Nibea albifora (Perciformes, Sciaenidae). Mitochondrial DNA. 22:80–82. [DOI] [PubMed] [Google Scholar]
- Cheng YZ, Xu TJ, Shi G, Wang RX. 2010. Complete mitochondrial genome of the miiuy croaker Miichthys miiuy (Perciformes, Sciaenidae) with phylogenetic consideration. Mar Genomics. 3:201–209. [DOI] [PubMed] [Google Scholar]
- Jin XX, Wang RX, Xu TJ, Shi G. 2012. Complete mitochondrial genome of Oxuderces dentatus (Perciformes, Gobioidei). Mitochondrial DNA. 23:142–144. [DOI] [PubMed] [Google Scholar]
- Lavoue S, Miya M, Nishida M. 2010. Mitochondrial phylogenomics of anchovies (family Engraulidae) and recurrent origins of pronounced miniaturization in the order Clupeiformes. Mol Phylogenet Evol. 56:480–485. [DOI] [PubMed] [Google Scholar]
- Li Q, Wu X, Shu H, Yang H, Yang L, Yue L. 2014The complete mitochondrial genome of the Gray's grenadier anchovy Coilia grayii (Teleostei, Engraulidae). Mitochondrial DNA. 27:175–176 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual. Cold spring Harbor (NY): CSH Laboratory Press. [Google Scholar]
- Shi G, Jin XX, Zhao SL, Xu TJ, Wang RX. 2012. Complete mitochondrial genome of Trypauchen vagina (Perciformes, Gobioidei). Mitochondrial DNA. 23:151–153. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu TJ, Cheng YZ, Sun YN. 2011. The complete mitochondrial genome of bighead croaker, Collichthys niveatus (Perciformes, Sciaenidae): structure of control region and phylogenetic considerations. Mol Biol Rep. 38:4673–4685. [DOI] [PubMed] [Google Scholar]
- Zhang B, Sun Y. 2013. Complete mitochondrial genome of Setipinna taty (Scaly hair-fin anchovy): repetitive sequences in the control region. Mitochondrial DNA. 24:616–618. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xian W. 2014. The complete mitochondrial genome of the larvae Osbeck's grenadier anchovy Coilia mystus (Clupeiformes, Engraulidae) from Yangtze estuary. Mitochondrial DNA. 27:966–967. [DOI] [PubMed] [Google Scholar]