Abstract
The complete mitochondrial genome of the edible fungus Pleurotus eryngii (oyster mushroom) was determined using Illumina sequencing. This mitogenome is a circular molecule of 72,650 bp in length with a GC content of 26.28%. Gene prediction showed that 40 putative protein-coding genes, the small ribosomal RNA subunits (rns), and 23 tRNAs were located on the same strand. The mitogenome of P. eryngii has a similar structure to that of P. ostreatus in both gene content and gene order. The mitogenome information of P. eryngii should contribute to our understanding of the diversity and evolution of Pleurotaceae and Agaricales.
Keywords: Pleurotus eryngii, agaricales, pleurotaceae, mitochondrial genome
Pleurotus species are globally distributed macrofungi and contain several well-known cultivated edible mushrooms. These fungi can secrete a diversity of extracellular enzymes (cellulases, hemicellulases, pectinases, ligninases, proteases, peptidases and so on) that can break down complex plant biomolecules, including cellulose, hemicellulose and lignin (Cohen et al. 2002; Yao & Jin 2004; Xie et al. 2016). As such, they play an essential role in nutrient cycling in natural environments. Pleurotus eryngii is an edible mushroom with a high commercial value and has been widely cultivated in Europe, Middle East, North America, and Asia (Mandeel et al. 2005). The genetic diversity and population structure of P. eryngii have been investigated in several studies (Urbanelli et al. 2003, 2007; Abdollahzadeh et al. 2007; Ro et al. 2007; Zervakis et al. 2014). However, little is known about the mitochondrial genome of this fungus. In this study, we report the complete mitogenome sequence of P. eryngii (KX827267) and provide a phylogenetic analysis of related taxa based on concatenated mitochondrial protein-coding genes.
The monokaryotic strain of P. eryngii 181 was isolated from a dikaryotic strain ‘Xinghan’ using a protoplast isolation method published previously (Chang et al. 1985). Both strains are deposited in the Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences. Total DNA extraction, library construction, and Illumina sequencing were performed according to the methods by Karlsson et al. (2015). A total of 22,476,346 and 14,370,163 clean pair-end reads were generated from two libraries with insert sizes of 300 bp and 8000 bp, respectively. The reads were assembled using abyss (BC, Canada, Simpson et al. 2009). The mitochondrial genome of P. ostreatus (NC_003388) was served as the reference to extract the mitochondrial genome from the assembled scaffolds using BLAST (PA). After removing repetitive ends, the scaffold was linked into a circular molecule.
The complete sequence of P. eryngii mitogenome was 72,650 bp in length with a GC content of 26.28%. The circular mitogenome encoded 40 putative protein-coding genes, the small ribosomal RNAs (rns), and 23 tRNAs. The 14 conserved protein-coding genes encoded the 7 subunits of NAD dehydrogenase (nad1-6 and nad4L genes), 3 cytochrome oxidases (cox1-3), apocytochrome b (cob) and 3 subunits of ATP synthase (atp6, apt 8 and apt 9). The 23 tRNA genes covered all 20 standard amino acids, with the following three having two tRNAs each: 2 trnL (trnL-uaa and trnL-uag), 2 trnR (trnR-ucg and trnR-ucu) and 2 trnS (trnS-gcu and trnS-uga) while the remaining amino acids were each represented by only one tRNA gene. There were 8 introns distributed in 2 protein-coding genes, i.e. cob (1 intron) and cox1 (7 introns). These introns mainly belong to group IB. Compared with P. ostreatus, the composition and order of the protein and rRNA genes are highly conserved.
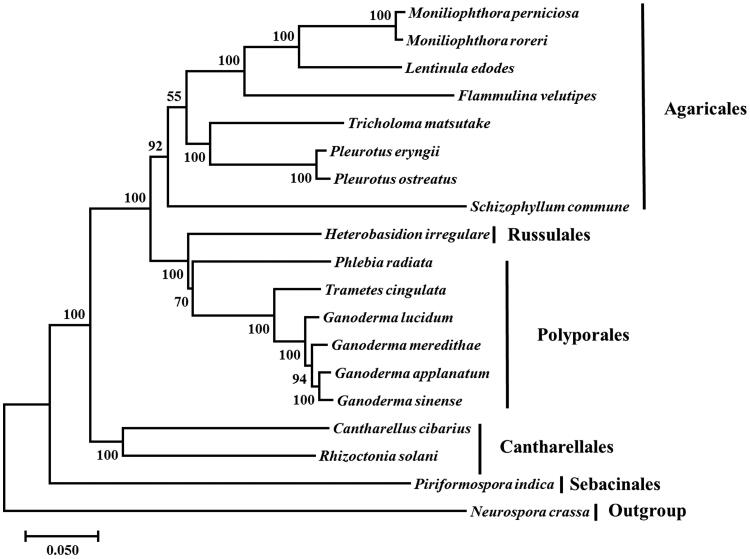
Phylogenetic analysis based on concatenated protein sequences confirmed that P. eryngii was a member of Agaricales and closely related to P. ostreatus. As shown in Figure 1, the monophyly of Pleurotaceae received strong support, which was in agreement with the monophyletic status of Pleurotaceae within the polyphyletic pleurotoid-lentinoid fungi as inferred based on nuclear gene sequences (Thorn et al. 2000). Our results also showed that the evolutionary relationships among Agaricales, Russulales, Polyporales, Cantharellales and Sebacinales based on mitochondrial genes were similar to results based on nuclear genes as revealed in previous studies (Hibbett 2006; Garciasandoval et al. 2011). The mitochondrial genome sequence of P. eryngii should help further studies of the diversity and evolution of Pleurotaceae and Agaricales.
Figure 1.
Phylogenetic analysis of 19 species of Agaricomycotina conducted by Neighbour-joining method as implemented in MEGA 7.0 (Tokyo, Japan, Kumar et al. 2016) based on concatenated amino acid sequences of 12 mitochondrial protein-coding genes, including atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4L, nad5 and nad6. The concatenated sequences were aligned using Clustal X (Thompson et al. 2010). The 19 species belonged to Agaricomycotina used for phylogenetics were listed below: Cantharellus cibarius (NC_020368), Flammulina velutipes (NC_021373), Ganoderma applanatum (NC_027188), Ganoderma lucidum (NC_021750), Ganoderma meredithae (NC_026782), Ganoderma sinense (NC_022933), Heterobasidion irregulare (NC_024555), Lentinula edodes (NC_018365), Moniliophthora perniciosa (NC_005927), Moniliophthora roreri (NC_015400), Phlebia radiata (NC_020148), Pleurotus ostreatus (NC_009905), Rhizoctonia solani (HF546977), Schizophyllum commune (NC_003049), Serendipita indica (FQ859090), Trametes cingulata (NC_013933) and Tricholoma matsutake (NC_028135). Neurospora crassa (NC_026614) was served as outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
References
- Abdollahzadeh J, Asef MR, Mirmahmoodi T.. 2007. The Pleurotus eryngii species-complex in Kurdistan region of Iran . Pak J Biol Sci. 10:3006–3009. [DOI] [PubMed] [Google Scholar]
- Chang ST, Li GSF, Peberdy JF.. 1985. Isolation of protoplasts from edible fungi. Mircen J Appl Microbiol Biotechnol. 1:185–193. [Google Scholar]
- Cohen R, Persky L, Hadar Y.. 2002. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus . Appl Microbiol Biotechnol. 58:582–594. [DOI] [PubMed] [Google Scholar]
- Garciasandoval R, Wang Z, Binder M, Hibbett DS.. 2011. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 103:510–524. [DOI] [PubMed] [Google Scholar]
- Hibbett DS. 2006. A phylogenetic overview of the Agaricomycotina. Revue Belge De Philologie Et Dhistoire. 10:917–925. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Durling MB, Choi J, Kosawang C, Lackner G, Tzelepis GD, Nygren K, Dubey MK, Kamou N, Levasseur A, et al. 2015. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol Evol. 7:465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeel QA, Al-Laith AA, Mohamed SA.. 2005. Cultivation of oyster mushrooms (Pleurotus spp.) on various lignocellulosic wastes. World J Microbiol Biotechnol. 21:601–607. [Google Scholar]
- Ro HS, Kim SS, Ryu JS, Jeon CO, Lee TS, Lee HS.. 2007. Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol Res. 111:710–715. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I.. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG.. 2010. The CLUSTAL_X windows interface: exible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn RG, Moncalvo JM, Reddy CA, Vilgalys R.. 2000. Phylogenetic analyses and the distribution of nematophagy support a monophyletic pleurotaceae within the polyphyletic pleurotoid-lentinoid fungi. Mycologia. 92:241–252. [Google Scholar]
- Urbanelli S, Della RV, Fanelli C, Fabbri AA, Reverberi M.. 2003. Genetic diversity and population structure of the Italian fungi belonging to the taxa Pleurotus eryngii (DC.:Fr.) Quel and P. ferulae (DC.:Fr.) Quel. Heredity. 90:253–259. [DOI] [PubMed] [Google Scholar]
- Urbanelli S, Rosa VD, Punelli F, Porretta D, Reverberi M, Fabbri AA, Fanelli C.. 2007. DNA-fingerprinting (AFLP and RFLP) for genotypic identification in species of the Pleurotus eryngii complex . Appl Microbiol Biotechnol. 74:592–600. [DOI] [PubMed] [Google Scholar]
- Xie C, Luo W, Li Z, Yan L, Zhu Z, Wang J, Hu Z, Peng Y.. 2016. Secretome analysis of Pleurotus eryngii reveals enzymatic composition for ramie stalk degradation. Electrophoresis. 37:310–320. [DOI] [PubMed] [Google Scholar]
- Yao Z, Jin L.. 2004. Advances in the research of Pleurotus eryngii. Acta Edulis Fungi. 11:52–58. [Google Scholar]
- Zervakis GI, Ntougias S, Gargano ML, Besi MI, Polemis E, Typas MA, Venturella G.. 2014. . A reappraisal of the Pleurotus eryngii complex - new species and taxonomic combinations based on the application of a polyphasic approach, and an identification key to Pleurotus taxa associated with Apiaceae plants. Fungal Biol. 118:814–834. [DOI] [PubMed] [Google Scholar]