Abstract
The horn-nosed boxfish, Ostracion rhinorhynchos (Tetraodontiformes: Ostraciidae) is a toxic marine species inhabiting tropical coral reefs. In this study, we first reported its whole mitochondrial genome sequence. The complete mitochondrial genome, 16 483 bp with an AT ratio of 56.8%, is composed of 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs and an 826-bp D-loop control region. The molecular-based phylogenetic tree indicated that O. rhinorhynchos has close affinities with fishes from family Ostraciidae as expected.
Keywords: Complete mitochondrial genome, Ostracion rhinorhynchos, tetraodontiformes
First described in 1851 (Bleeker 1851), Ostracion rhinorhynchos, commonly known as horn-nosed boxfish because of its box-like carapace and large protuberance on the snout, is a toxic marine genus belonging to Ostraciidae in Tetraodontiformes of which most species dwell in and around tropical coral reefs. Ostraciidae contains about 23 extant species in 6 extant genera, but mitochondrial genomes of only two species (Ostracion immaculatus and Lactoria diaphana) have been sequenced. In this study, we first reported the whole mitochondrial genome sequence of O. rhinorhynchos (GenBank accession number KU308378).
One adult horn-nosed boxfish was collected from Sanya, China. Whole genomic DNA was extracted from its muscle with Puregene Tissue Core Kit A (Qiagen, Germantown, MD, USA) and sequenced by Illumina Hiseq4000 (BGI, Shenzhen, China). Raw data containing adaptor contamination (with >15 bp matched to the adaptor sequence), polyNs (>5 bp Ns) or >1% error rate (>10 bp bases with quality score <20) were filtered out with a Perl script (Zhou et al. 2013; Tang et al. 2014). Clean reads were subsequently assembled with SOAPdenovo-Trans (Xie et al. 2014) and annotated with DOGMA (Wyman et al. 2004). tRNA genes were further identified using tRNAscan-SE 1.21 (http://lowelab.ucsc.edu/tRNAscan-SE).
The complete mitochondrial genome of O. rhinorhynchos is 16 483 bp in length. The overall base composition is 29.7% A, 27.1% T, 27.7% C and 15.4% G, with an AT bias of 56.8%, in common with other vertebrate mitochondrial genomes and slightly higher than that in reported O. immaculatus (Yamanoue et al. 2007). This circular molecule contains 13 protein-coding genes, 22 transfer RNA gens, 2 ribosomal RNA genes (12S rRNA and 16S rRNA) and an 826-bp D-loop control region (Table 1). ND6 is the only protein coding gene coded by L-strand while 14 out of 22 tRNAs are coded by H-strand. The 13 protein coding genes were aligned to two reported Ostraciidae fishes (O. immaculatus and L. diaphana) with Blastall (Mount 2007) and the result shows that, except for ATP8 which presents the same identity (93.94%), other 12 genes all have higher identity in O. immaculatus than those in L. diaphana, confirming that O. rhinorhynchos and O. immaculatus which are in the same genus have relatively closer phylogenetic relationship (Table 1).
Table 1.
Mitochondrial genome characteristics of the Ostracion rhinorhynchos.
| Gene name | Position |
Size (bp) | Intergenic nucleotidesa | Coding strand | Identity with Ostraciidae species (%) |
||
|---|---|---|---|---|---|---|---|
| Start | End | Ostracion immaculatus | Lactoria diaphana | ||||
| tRNA-Phe | 1 | 68 | 68 | 0 | H | / | / |
| s-rRNA | 69 | 1011 | 943 | 6 | H | / | / |
| tRNA-Val | 1018 | 1089 | 72 | 0 | H | / | / |
| l-rRNA | 1090 | 2771 | 1682 | 2 | H | / | / |
| tRNA-Leu | 2774 | 2847 | 74 | 0 | H | / | / |
| ND1 | 2848 | 3819 | 972 | 7 | H | 91.87 | 84.36 |
| tRNA-Ile | 3827 | 3897 | 71 | −1 | H | / | / |
| tRNA-Gln | 3897 | 3967 | 71 | −1 | L | / | / |
| tRNA-Met | 3967 | 4035 | 69 | 0 | H | / | / |
| ND2 | 4036 | 5079 | 1044 | 2 | H | 92.24 | 83.33 |
| tRNA-Trp | 5082 | 5153 | 72 | 0 | H | / | / |
| tRNA-Ala | 5154 | 5222 | 69 | 1 | L | / | / |
| tRNA-Asn | 5224 | 5296 | 73 | 37 | L | / | / |
| tRNA-Cys | 5334 | 5400 | 67 | 0 | L | / | / |
| tRNA-Tyr | 5401 | 5471 | 71 | 1 | L | / | / |
| COX1 | 5473 | 7020 | 1548 | 4 | H | 95.28 | 87.34 |
| tRNA-Ser | 7025 | 7095 | 71 | 3 | L | / | / |
| tRNA-Asp | 7099 | 7169 | 71 | 7 | H | / | / |
| COX2 | 7177 | 7866 | 690 | 1 | H | 96.52 | 89.57 |
| tRNA-Lys | 7868 | 7942 | 75 | 1 | H | / | / |
| ATP8 | 7944 | 8108 | 165 | −7 | H | 93.94 | 93.94 |
| ATP6 | 8102 | 8782 | 681 | 2 | H | 95.15 | 86.93 |
| COX3 | 8785 | 9567 | 783 | 2 | H | 95.79 | 88.89 |
| tRNA-Gly | 9570 | 9641 | 72 | 0 | H | / | / |
| ND3 | 9642 | 9989 | 348 | 1 | H | 92.24 | 81.9 |
| tRNA-Arg | 9991 | 10 060 | 70 | 0 | H | / | / |
| ND4L | 10 061 | 10 354 | 294 | −4 | H | 97.28 | 87.41 |
| ND4 | 10 351 | 11 730 | 1380 | 1 | H | 93.81 | 84.75 |
| tRNA-His | 11 732 | 11 800 | 69 | 0 | H | / | / |
| tRNA-Ser | 11 801 | 11 868 | 68 | 4 | H | / | / |
| tRNA-Leu | 11 873 | 11 945 | 73 | 0 | H | / | / |
| ND5 | 11 946 | 13 781 | 1836 | 2 | H | 93.03 | 85.19 |
| ND6 | 13 784 | 14 302 | 519 | 0 | L | 91.71 | 84.39 |
| tRNA-Glu | 14 303 | 14 371 | 69 | 4 | L | / | / |
| CYTB | 14 376 | 15 515 | 1140 | 1 | H | 92.89 | 84.7 |
| tRNA-Thr | 15 517 | 15 588 | 72 | −1 | H | / | / |
| tRNA-Pro | 15 588 | 15 657 | 70 | 0 | L | / | / |
| D-loop | 15 658 | 16 483 | 826 | 0 | / | / | |
Positive numbers indicate the number of nucleotides found in intergenic spacers between different genes. Negative numbers indicate overlapping nucleotides between adjacent genes.
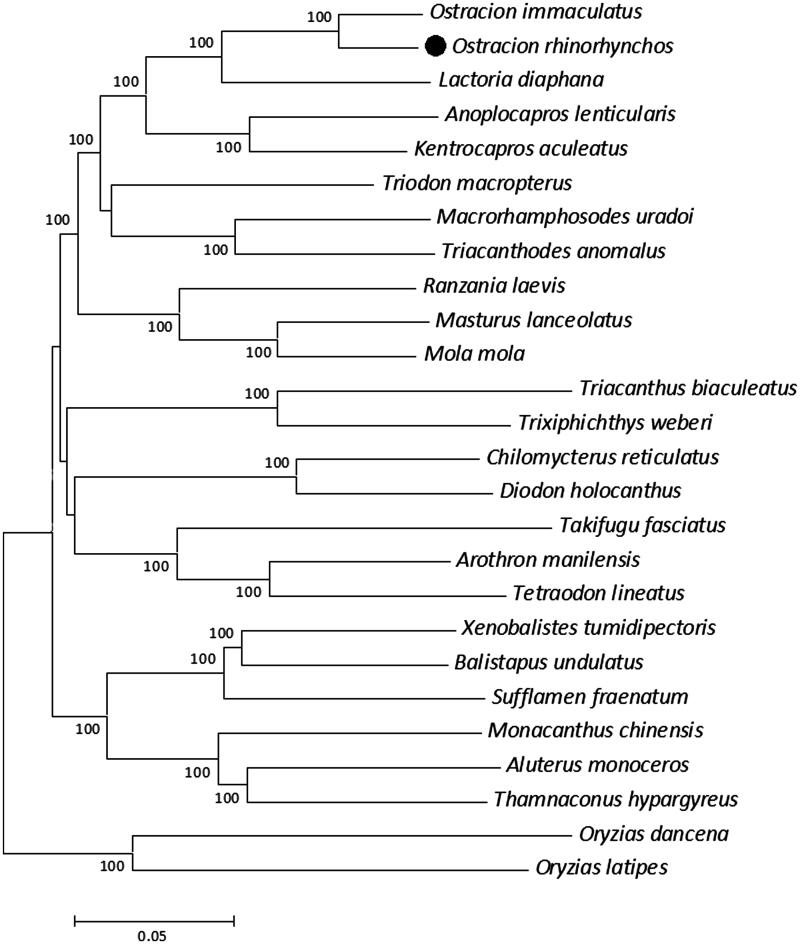
Several studies have confirmed the phylogenetic position of Tetraodontiformes fishes as a monophyletic group within the higher teleosts (Holcroft 2004; Yamanoue et al. 2007). However, within this order, the genetic relationship is not clear (Yamanoue et al. 2007; Santini et al. 2013). Taking Oryzias latipes and Oryzias dancena in order Beloniformes as outgroups, we constructed a neighbour-joining tree using MEGA6 (Tamura et al. 2013), based on the complete mitochondrial genomes of O. rhinorhynchos together with other 23 reported affinis species in order Tetraodontiformes (Figure 1). As expected, O. rhinorhynchos has closer affinities with other Ostraciidae species. The achieved mitochondrial genome of O. rhinorhynchos will be useful for verification of the evolutionary relationship within Tetraodontiformes.
Figure 1.
Phylogenetic tree based on Ostracion rhinorhynchos together with other 25 reported species. GenBank accession numbers of mitochondrial genome sequences are listed as follows: Aluterus monoceros: NC_027268.1; Anoplocapros lenticularis: NC_011319.1; Arothron manilensis: NC_015371.1; Balistapus undulatus: NC_011946.1; Chilomycterus reticulatus: NC_011331.1; Diodon holocanthus: NC_009866.1; Kentrocapros aculeatus: NC_009864.1; Lactoria diaphana: NC_011330.1; Macrorhamphosodes uradoi: NC_009860.1; Masturus lanceolatus: NC_005837.1; Mola mola: NC_005836.1; Monacanthus chinensis: NC_011925.1; Oryzias dancena: NC_012976.1; Oryzias latipes: NC_004387.1; Ostracion immaculatus: NC_009865.1; Ranzania laevis: NC_007887.1; Sufflamen fraenatum: NC_004416.1; Takifugu fasciatus: NC_013087.1; Tetraodon lineatus: NC_028551.1; Thamnaconus hypargyreus: NC_027070.1; Triacanthodes anomalus: NC_009861.1; Triacanthus biaculeatus: NC_009863.1; Triodon macropterus: NC_009859.1; Trixiphichthys weberi: NC_009862.1; Xenobalistes tumidipectoris: NC_011321.1.
Acknowledgements
We would like to thank Chuanyu Guo from BGI, Shenzhen for collecting precious samples for this study.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing. .
Funding information
This work was supported by Three New Projects of Agricultural Aquaculture Program of Jiangsu Province (No. Y2015-12), Special Project on the Integration of Industry, Education and Research of Guangdong Province (No. 2013B090800017), Shenzhen and Hong Kong Innovation Circle (No. SGLH20131010105856414) and Fish-T1K (Transcriptomes for 1000 Fishes) project (www.fisht1k.org).
References
- Bleeker P. 1851. Bijdrage tot de kennis der Balistini en Ostraciones van den Indischen Archipel. Verb Bat Gen. 24:1–38. [Google Scholar]
- Holcroft NI. 2004. A molecular test of alternative hypotheses of tetraodontiform (Acanthomorpha: Tetraodontiformes) sister group relationships using data from the RAG1 gene. Mol Phylogenet Evol. 32:749–760. [DOI] [PubMed] [Google Scholar]
- Mount DW. 2007. Using the basic local alignment search tool (BLAST). CSH Protoc. 1(2007):pdb.top17. [DOI] [PubMed] [Google Scholar]
- Santini F, Sorenson L, Marcroft T, Dornburg A, Alfaro ME.. 2013. A multilocus molecular phylogeny of boxfishes (Aracanidae, Ostraciidae; Tetraodontiformes). Mol Phylogenet Evol. 66:153–160. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, et al. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30:1660–1666. [DOI] [PubMed] [Google Scholar]
- Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M.. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol Phylogenet Evol. 45:89–101. [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Y, Liu S, Yang Q, Su X, Zhou L, Tang M, Fu R, Li J, Huang Q.. 2013. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. GigaScience 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]