Abstract
Trident maple Acer buergerianum Miq., belonging to the family of Aceraceae, is an important ornamental tree. Here, we report the complete chloroplast genome sequence of A. buergerianum. The circular genome was 156,477 bp in size, and comprised a pair of inverted repeat (IR) regions of 26,090 bp, a large single-copy (LSC) region of 86,246 bp and a small single-copy (SSC) region of 18,062 bp. It contained 134 genes, including 89 protein-coding genes, 40 transfer RNA genes, and 8 ribosomal genes. Maximum likelihood phylogenomic analysis shows that A. buergerianum is closely related to other Acer species, including A. miaotaiense, A. morrisonense, and A. davidii.
Keywords: Chloroplast genome, Acer buergerianum, phylogenomic analyses
Trident maple Acer buergerianum Miq., a member of the family of Aceraceae, is native to eastern China (Hayashi 2007) and Japan (Matsue & Takeda 2009). As an important ornamental tree, it has enormous commerce value. In recent decades human activities have seriously destroyed natural habitats, leading to the loss and quick decline of a large number of natural populations of wild trident maple in China. As plant chloroplast sequences can provide helpful information to reconstruct complex evolutionary relationships, they have been widely applied for species identification and phylogenetic analysis during the past years (Jansen et al. 2007; Moore et al. 2007; Parks et al. 2009; Ruhsam et al. 2015). The information of chloroplast genomes has been extensively applied in understanding plant genetic diversity and conservation genetics (Ye et al. 2014; Zhang et al. 2016). Thus, the obtainability of chloroplast genome of A. buergerianum will prominently advance the conservation efforts of this threatened plant species that harbours valuable gene sources for the breeding program of trident maple.
In this study, we sequenced, assembled, and annotated the chloroplast genome of A. buergerianum using high-throughput genome sequencing technology. Young, fresh and healthy leaves of A. buergerianum were collected from a wild trident maple tree from Xupu, Hunan Province, China (110.6086E, 27.9101N). Total genomic DNA was extracted using a modified CTAB method (Doyle & Doyle 1987). According to the Illumina’s protocol, the pair-end libraries were constructed and then sequenced using Hiseq2000 platform, obtaining ∼50.1 million high-quality clean reads. With A. miaotaiense (Zhang et al. 2016) as a reference, we assembled and annotated the cp genome using CLC Genomic Workbench v7.5 (CLC BIO, Denmark) and Dogma (https://dogma.ccbb.utexas.edu/), respectively. The annotated genomic sequence was deposited into the GenBank database under the accession number of KY419137. The complete chloroplast genome is 156,477 bp in size, containing a large single copy region (LSC) of 86,246 bp, a small single copy region (SSC) of 18,062bp, and a pair of 26,090 bp inverted repeat regions (IRs). The chloroplast genome harbours 134 genes, including 89 protein-coding genes, 40 tRNA genes and 8 rRNA genes. The base composition is asymmetric (A: 30.65%; C: 19.30%; G: 18.58%; T: 31.47) with an overall GC content of 37.88%.
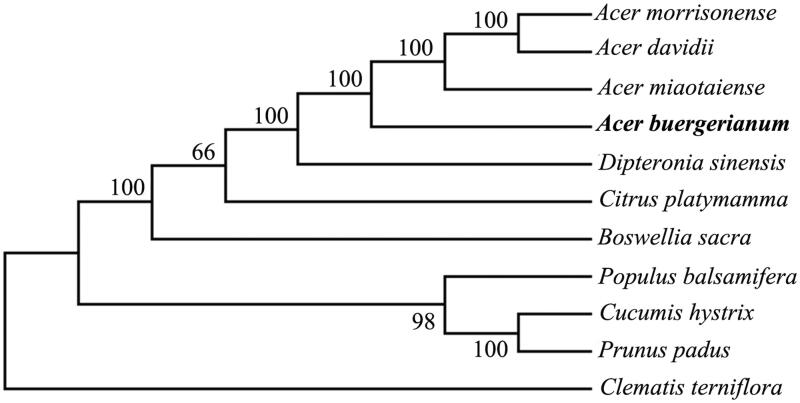
To determine the phylogenetic position of A. buergerianum, we constructed the phylogenetic tree between A. buergerianum and 10 other plant species based on whole chloroplast genome sequences with maximum likelihood (ML) method using MEGA7 (Kumar et al. 2016) (Figure 1). As expected, A. buergerianum is closely related to other Acer species, including A. miaotaiense, A. morrisonense, and A. davidii, and they form a clade together with Dipteronia sinensis belonging to the family Aceraceae with a strong bootstrap value of 100%. The complete chloroplast genome of A. buergerianum will provide valuable genetic data for the conservation management of the precious ornamental tree species.
Figure 1.
Maximum likelihood phylogenetic tree of A. buergerianum with 10 other plant species based on complete chloroplast genome sequences using Clematis terniflora as outgroup. Numbers in the nodes are bootstrap values from 1000 replicates. Accession numbers are listed as below: Acer morrisonense NC_029371.1, Acer davidii NC_030331.1, Acer miaotaiense NC_030343.1, Dipteronia sinensis NC_029338.1, Citrus platymamma NC_030194.1, Boswellia sacra NC_029420.1, Populus balsamifera NC_024735.1, Cucumis hystrix NC_023544.1, Prunus padus NC_026982.1, Clematis terniflora NC 028000.1.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hayashi M. (2007). Tree bark handbook. Tokyo: Bun-ichi Co., Ltd. [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, et al. . 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci. 104:19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsue M, Takeda Y. (2009). Technical Note of National Institute for Land and Infrastructure Management, No. 506. The Street Tree of Japan VI, National Institute for Land and Infrastructure Management, Ministry of Land, Infrastructure, Transport, and Tourism; p. 76. [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE.. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci. 104:19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A.. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Ennos RA.. 2015. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Res. 15:1067–1078. [DOI] [PubMed] [Google Scholar]
- Ye CY, Lin Z, Li G, Wang YY, Qiu J, Fu F, Zhang H, Chen L, Ye S, Song W.. 2014. Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PLoS One. 9:e113657-e113657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li K, Gao J, Liu Y, Gao LZ.. 2016. The complete plastid genome sequence of the wild rice Zizania latifolia and comparative chloroplast genomics of the rice Tribe Oryzeae, Poaceae. Front Ecol Evol. 4:88. [Google Scholar]
- Zhang Y, Li B, Chen H, Wang Y.. 2016. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conserv Genet Res. 8:1–3. [Google Scholar]