Abstract
The complete mitochondrial genome sequence of Chinese native Dolly Varden (Salvelinus malma sp.) was sequenced. It was 16,652-nucleotide in length and consisted of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 non-coding regions (L-strand replication origin and control region), showing conserved gene arrangement with most vertebrates. The phylogenetic analysis based on Cytb genes of Chinese native as well as four other Dolly Varden subspecies showed that Chinese native S. malma sp. had close relationships with S. curilus (formerly named as S. malma krascheninnikova) and S. malma miyabe. This result did not support the monophyletic relationship of Dolly Varden chars. Further studies are needed to determine the phylogenetic position of Chinese native Dolly Varden.
Keywords: Salvelinus malma sp.;· mitochondrion;· genome
Dolly Varden is a large anadromous or freshwater char (Salvelinus malma). It inhabits in the streams of north-western North America, Russian Far East, Korea, Japan, as well as in Northeast of China (Armstrong and Morrow 1980). However, little genetic information of Chinese native Dolly Varden (Salvelinus malma sp.) is available. In this study, we sequenced and described the complete mitochondrial genome of S. malma sp. native in China for the first time.
The specimen was collected from the upper Yalu River in Jilin Province, China. The samples were stored in −80 °C in Research Institute of Marine Biotechnology and life Health, Ningbo University, Ningbo. Total DNA was extracted from dorsal fin following TIANamp Marine Animals DNA Kit (Tiangen, China). Totally 28 pairs of primers were used, in which 16 pairs of primers were retrieved from Gonostoma gracile (Miya and Nishida 1999) and the remaining primers were designed based on public mitochondrial genome sequences of Salvelinus albus (white char), S. fontinalis and S. curilus (with accession number of NC_028018, NC_000860, NC_024585, respectively).
The sequenced fragments were de novo assembled into complete mitogenome and annotated by comparing with published genome sequences of other vertebrate species using Mega5.1 (Tamura et al. 2011) and MitoFish (Iwasaki et al. 2013). Finally, a physical map of S. malma sp. mitogenome was generated and uploaded to GenBank with accession number MF680544.
The complete mitogenome of Chinese native Dolly Varden was 16,652 bp in length. The genomic organization was identical to those of typical vertebrate mitochondrial genomes, including two rRNA genes, 13 protein-coding genes, 22 tRNA genes, a light-strand replication origin (OL), and a putative control region (CR). The overall base composition was 28.1% of A, 26.4% of T, 28.5% of C, and 17.0% of G with a slight A + T bias (54.5%) like other vertebrate mitochondrial genomes. The 13 protein-coding genes totally encoded 3808 amino acids and their base composition was 25.7%, 9.0%, 29.0%, and 16.3% for A, T, C, and G, respectively. The features mentioned above were accordant with typical Salvelinus fish mitogenome (Salmenkova et al. 2009; Balakirev et al. 2016).
For the 13 protein-coding genes, 12 genes started with ATG while only COI started with GTG. Six genes shared the termination codon TAA (COI, ATPase8, ND1, ND2, ND5, and ND4L), one with TAG (ND6), the remaining with incomplete stop codon (COII, COIII, ND3, ND4, ATPase6, and Cytb). This feature was common among vertebrate mitochondrial protein-coding genes (Cheng et al. 2010). Chinese native S. malma sp. had two non-coding regions, the L-strand replication origin region (36 bp) located between tRNA-Asn and tRNA-Cys, and the control region located within the tRNA-Pro and tRNA-Phe. The 13 protein-encoding genes of Chinese native S. malma sp. mitochondrion encoded 3808 amino acids with stop codons included. Leucine was the most frequent amino acid while cystine was the least frequent. CTT was the most frequent used codon (4.46%) and AAG was the least (0.11%), showing similar features with those of other vertebrates (Miya et al. 2003).
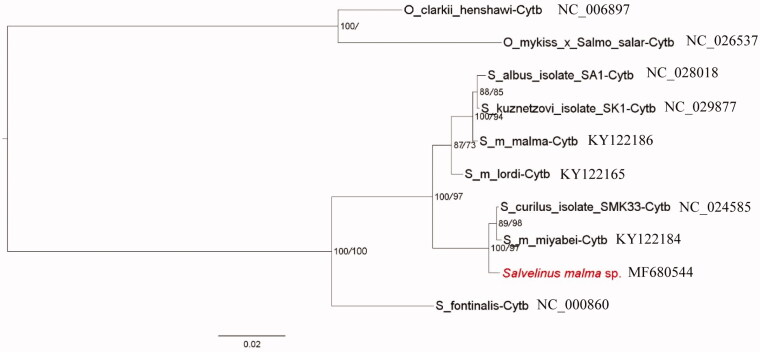
To explore the phylogenetic position of Chinese native Dolly Varden, phylogenies based on maximum likelihood (ML) and Bayesian method were constructed using public Cytb genes involving other nine Salmonidae species. Two species, Oncorhynchus clarkii henshawi and O. mykiss × Salmo salar, were used as outgroups. The nucleotide sequences were aligned by ClustalX with default settings and calculated the best-fit nucleotide model (TrN + I) (Tamura and Nei 1993) by jModelTest 0.1 (Posada 2008). Then the ML and Bayesian analyses were conducted by PhyML3.1 (Guindon et al. 2010) and MrBayes (Huelsenbeck and Ronquist 2001) and validated by 1000 bootstraps and posterior probability, respectively. The ML and Bayesian methods resulted in the same phylogeny, showing that Chinese native Dolly Varden (S. malma sp.) had a close relationship with S. curilus (a.k.a. S. malma krascheninnikova) and S. malma miyabe (Figure 1), forming an Asian group while S. malma lordi and S. malma malma, together with S. albus and S. kuznetzovi forming another group. It did not support the monophyletic relationship of S. malma fish. Further investigations on morphological and molecular comparisons are needed to resolve the phylogenetic position of Chinese native Dolly Varden.
Figure 1.
Phylogenetic tree of Dolly Varden chars as inferred from the nucleotide sequences of partial Cytb genes. The phylogeny was reconstructed by maximum likelihood and Bayesian method, respectively. The percent values of bootstrap/posterior probability were showed at each node. And the Chinese Dolly Varden was shown in red.
Disclosure statement
The authors report no confiicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Armstrong RH, Morrow J.. 1980. The Dolly Varden charr: Salvelinus malma. New York (NY): Springer. [Google Scholar]
- Balakirev ES, Romanov NS, Ayala FJ.. 2016. Complete mitochondrial genomes of the Northern (Salvelinus malma) and Southern (Salvelinus curilus) Dolly Varden chars (Salmoniformes, Salmonidae). Mitochondrial DNA A. 27:1016–1017. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Shi G, Wang R.. 2010. Complete mitochondrial genome of the miiuy croaker Miichthys miiuy (Perciformes, Sciaenidae) with phylogenetic consideration. Marine Genomics. 3:201–209. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755. [DOI] [PubMed] [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M.. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya M, Nishida M.. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei:Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426. [DOI] [PubMed] [Google Scholar]
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. . 2003. Major patterns of higher teleostean phylogenies:a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138. [DOI] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Salmenkova EA, Omel’chenko VT, Afanasyev KI, Rubtsova GA, Kovalev MY.. 2009. Genetic divergence of populations of the white char Salvelinus albus, northern and southern forms of malma S. malma (Salmonidae), by microsatellite loci of DNA. J Ichthyol. 49:730–740. [Google Scholar]
- Tamura K, Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]