Abstract
The paper mulberry, Broussonetia kazinoki × Broussonetia papyifera, is a species within the family Moraceae, also is an economic and ecological plant. Complete chloroplast genome of paper mulberry was reported in this study. The total genome size was 160,903 bp in length, which is separated by a large single copy (LSC) region and a small single copy (SSC) region (89,220 and 20,079 bp in length, respectively). There were a total of 134 predicted functional genes, including 80 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 46 transfer RNA (tRNA) genes. The overall G + C content of the paper mulberry chloroplast genome was 35.74%, while the corresponding values of the LSC, SSC, and IR regions are 33.33%, 28.50%, and 42.72%, respectively. The phylogenetic analysis between Moraceae trees showed chloroplast genome of paper mulberry was closely related to Morus multicaulis.
Keywords: Paper mulberry, chloroplast genome, Illumina sequencing, Moraceae
The paper mulberry, hybridized by Broussonetia kazinoki × Broussonetia papyifera, is a species of Moraceae, which is distributed in Eastern Asia and pacific countries (Xianjun et al. 2014). The species has important economic values and ecological values. The bark is often used to make paper and cloth (Saito et al. 2009). The leaves have been used as a kind of forage for livestock due to the high contents of protein. The abundant flavonoids in paper mulberry have long been used in Chinese medicine (Sun et al. 2012). In terms of ecological values, the tree grow fast and have strong tolerance to cold, drought and salt (Li et al. 2011; Sun et al. 2014; Peng et al. 2015), which can be planted as a pioneer tree in ecological degradation area. Thus, it is useful to know more genetic information about paper mulberry to reveal the mechanism of evolution or adoption.
The whole genomic DNA was extracted from fresh leaves of paper mulberry seedlings with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). The seedlings were collected from Yuexi, China (115°55′–116°33′E, 30°39′–31°11′N). The specimen was kept in the laboratory at −80 °C under the accession number 20170520GS. DNA was shared by Covaris M220 (Covaris, Woburn, MA), yielding fragments of 500 bp in length. Then, an Illumina pair-endlibrary was constructed and sequenced by the Illumina HiSeq 4000 platform. After filtered out the low-quality reads by Trimmomatic V0.32 (Bolger et al. 2014), the clean reads were used for the chloroplast genome assembly by velvet v1.2.07 (Zerbino and Birney 2008). Sequences were annotated in DOGMA (Wyman et al. 2004). A circular map of the plastome was generated using OGDRAW (Lohse et al. 2007). The complete chloroplast sequence of paper mulberry has been submitted to GenBank with the accession number of MF496038.
The complete chloroplast genome of paper mulberry is 160,903 bp in length, comprising a large single copy (LSC) region of 89,220 bp and a small single copy (SSC) region of 20,079 bp, separated by two inverted repeat regions (IRs) of 25,802 bp. The overall G + C content of chloroplast genome is 35.74%, while the corresponding values of the LSC, SSC, and IRs regions are 33.33%, 28.50%, and 42.72%, respectively. The chloroplast genome contains a total of 134 predicted functional genes, including 80 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 46 transfer RNA (tRNA) genes. Among the annotated genes, nine of them (atpF, rpoC1, rpl2 × 2, ndhB × 2, ndhA, ycf1, and trnL-TAA) contain one intron, and two genes (ycf3 and clpP) include two introns.
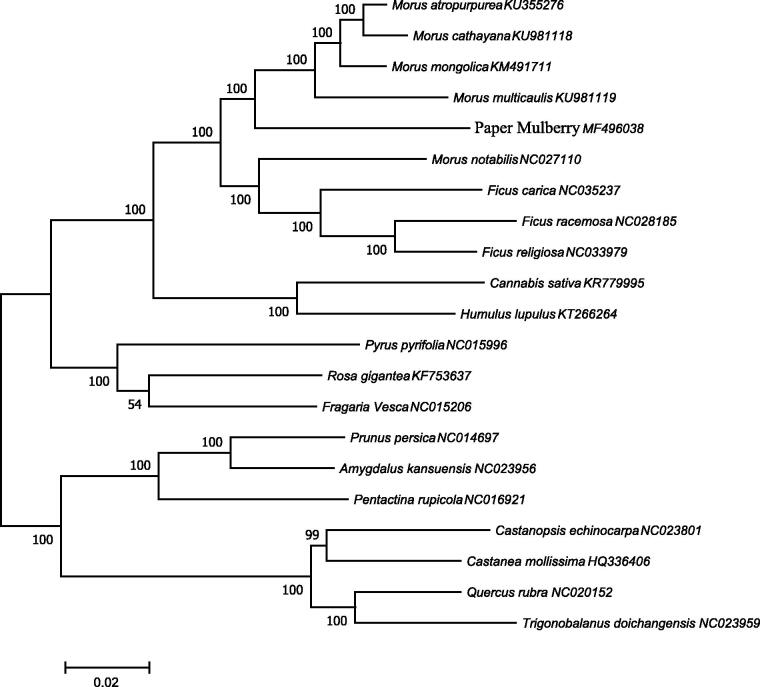
To ascertain phylogenetic position of paper mulberry, 21 complete chloroplast genomes sequences of Moraceae family were employed and phylogenetic analysis showed that paper mulberry was clustered into Morus and is closely related to Morus mongolica (Figure 1). The complete chloroplast genome information reported here provides a useful tool not only for high quality gene mining of paper mulberry, but also for phylogenetic and evolutionary studies within Moraceae.
Figure 1.
The phylogenetic tree based on 21 complete chloroplast genome sequences of Moraceae family. The neighbour-joining (NJ) phylogenetic tree was constructed with MEGA 7 (with 1000 bootstrap replicates) using 80 protein-coding genes of 21 species of Moraceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li Y, Li H, Wu G.. 2011. Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiol. 31:349–357. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R.. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Peng X, Wu Q, Teng L, Feng T, Zhi P, Shen S.. 2015. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 15:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Linquist B, Keobualapha B, Shiraiwa T, Horie T.. 2009. Broussonetia papyrifera (paper mulberry): its growth, yield and potential as a fallow crop in slash-and-burn upland rice system of northern Laos. Agroforest Syst. 76:525–532. [Google Scholar]
- Sun J, Liu SF, Zhang CS, Yu LN, Bi J, Zhu F, Yang QL.. 2012. Chemical composition and antioxidant activities of Broussonetia papyrifera fruits. PLoS One. 7:e32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Peng X, Fan W, Tang M, Liu J, Shen S.. 2014. Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene. 535:140–149. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xianjun P, Linhong T, Xiaoman W, Yucheng W, Shihua S.. 2014. De novo assembly of expressed transcripts and global transcriptomic analysis from seedlings of the paper mulberry (Broussonetia kazinoki × Broussonetia papyifera). PLoS One. 9:e97487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]