Abstract
The complete mitochondrial genome of Haemaphysalis concinna is reported for the first time in this study. Its entire mitogenome is 14,675 bp in length, contained 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and two non-coding regions. Among the 13 protein-coding genes, apart from the nad1, nad4, nad4L, and nad5 gene encoded on the L-strand, the remaining protein-coding genes were encoded on the H-strand. The phylogenetic analysis by Bayesian inference method shows that Amblyomma sphenodonti and H. parva formed one clade, while H. concinna and other species of genus Haemaphysalis formed the other clade, indicating that H. concinna belong to the genus Haemaphysalis.
Keywords: Haemaphysalis concinna, mitochondrial genome, phylogenetic analysis
The obligate haematophagous ectoparasites Haemaphysalis concinna (Ixodida: Ixodidae) is widely distributed in China (Teng and Jiang 1991), Russia, Germany, as well as temperate Eurasia (Nosek 1971). These adult stages of H. concinna mainly parasitize on Artiodactyla, and accidently attack humans that cause mechanical damage and transmit a great variety of pathogens (Mikryukova et al. 2014; Švehlová et al. 2014).
The adult of H. concinna was collected by swiping flags on vegetations from Seven peak national Forest Park (46.711100 N, 130.945300 E, 550 m at attitude), Huanan County Heilongjiang Province in Northeast China, on 12 April 2016. The individual tick was stored in the Department of Parasitology, Heilongjiang Bayi Agricultural University (specimen no. BYNKPL-160412), and DNA was extracted by TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), and stored at −20 °C until use. The entire mitochondrial genomic sequences were composed of two overlapping fragments, one fragment approximately 5.9 kb was amplified from cox1 to rrnL using the primers CO1-J (5′-CCT GAT ATA GCA TT TCC TCG-3′) and 16S-N (5′-CTG CTC AAT GAT TTT TAA ATT GCT GTG-3′), the other one fragment approximately 9.0 kb was amplified from rrnL to cox1 using the primers 16S-J (5′-TTA CGC TGT TAT CCC TAG AGT ATT-3′) and CO1-N (5′-GCT ATA TCA GGT GCA CCT-3′).
The entire H. concinna mt genome was a typical circular DNA molecule with 14,675 bp in size (GenBank accession number NC_034785), which contained 13 protein-coding genes (cox1-3, nad1-6, nad4L, atp6, atp8, and cytb), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two non-coding regions (NCRs). The protein-coding genes were transcribed in different directions, which were consistent with those of other ticks (Black and Roehrdanz 1998; Burger et al. 2014; Guo et al. 2016), but distinct from those of trematodes, cestodes, and nematodes (Yamasaki et al. 2012; Duan et al. 2015; Chang et al. 2016), which transcribed in the same directions. Among the 13 protein-coding genes, apart from the nad1, nad4, nad4L, and nad5 gene encoded on the L-strand, the remaining protein-coding genes were encoded on the H-strand. The nucleotide compositions of the complete mtDNA sequence of H. concinna were biased towards A + T (77.96%), with T being the most favoured nucleotide (39.35%) and G was the least favoured (9.21%). The H. concinna mt genome encoded 3615 amino acids in total. The A + T content of protein-coding genes ranged from 71.02% (cox1) to 84.57% (atp8).
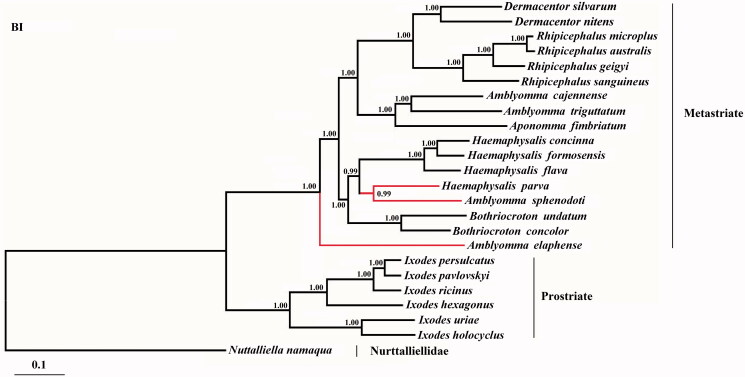
Based on the concatenated amino acid sequence dataset (13 protein-coding genes), phylogenetic analyses were performed using Bayesian inference (BI). The result showed that the tree was divided into two large branches: Prostriata and Metastriata (Figure 1). Within the metastriate, H. concinna and genus Haemaphysalis species clustered together with high statistical support (PP = 1), indicating that H. concinna belong to the genus Haemaphysalis. This study provides not only new mtDNA resource for phylogenetic studies, but also novel and useful genetic marker for further studies on species identification, population genetics, and molecular epidemiology of the genus Haemaphysalis in ticks.
Figure 1.
Phylogenetic relationships of Haemaphysalis concinna and other species based on mitochondrial sequence data. The concatenated amino acid sequences of 13 protein-coding genes were analysed with Bayesian inference (BI), using Nuttalliellidae namaqua (NC_019663) as an outgroup. All the species accession numbers in this study are listed as below: Amblyomma triguttatum NC_005963, Amblyomma elaphense NC_017758, Amblyomma sphenodonti NC_017745, Amblyomma cajennense NC_020333, Aponomma fimbriatum NC_017759, Bothriocroton concolor NC_017756, Bothriocroton undatum NC_017757, Haemaphysalis flava NC_005292, Haemaphysalis formosensis NC_020334, Haemaphysalis parva NC_020335, Rhipicephalus sanguineus NC_002074, Rhipicephalus microplus KP143546, Rhipicephalus australis NC_023348, Rhipicephalus geigyi NC_023350, Ixodes hexagonus NC_002010, Ixodes holocyclus NC_005293, Ixodes persulcatus NC_004370, Ixodes uriae NC_006078, Ixodes pavlovskyi NC_023831, Ixodes ricinus NC_018369, Dermacentor nitens NC_023349, Dermacentor silvarum NC_026552, and Nuttalliella namaqua NC_019663.
Disclosure statement
The authors declare no conflict of interest.
References
- Black WC, Roehrdanz RL.. 1998. Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol Biol Evol. 15:1772–1785. [DOI] [PubMed] [Google Scholar]
- Burger TD, Shao R, Barker SC.. 2014. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet Evol. 76:241–253. [DOI] [PubMed] [Google Scholar]
- Chang QC, Liu GH, Gao JF, Zhang X, Zhang Y, Duan H, Yue DM, Fu X, Su X, Wang CR.. 2016. Sequencing and characterization of the complete mitochondrial genome from the pancreatic fluke Eurytrema pancreaticum (Trematoda: Dicrocoeliidae). Gene. 576:160–165. [DOI] [PubMed] [Google Scholar]
- Duan H, Gao JF, Hou MR, Zhang Y, Liu ZX, Gao DZ, Guo DH, Yue DM, Su X, Fu X, et al. 2015. Complete mitochondrial genome of an equine intestinal parasite, Triodontophorus brevicauda (Chromadorea: Strongylidae): the first characterization within the genus. Parasitol Int. 64:429–434. [DOI] [PubMed] [Google Scholar]
- Guo DH, Zhang Y, Fu X, Gao Y, Liu YT, Qiu JH, Chang QC, Wang CR.. 2016. Complete mitochondrial genomes of Dermacentor silvarum and comparative analyses with another hard tick Dermacentor nitens. Exp Parasitol. 169:22–27. [DOI] [PubMed] [Google Scholar]
- Mikryukova TP, Moskvitina NS, Kononova YV, Korobitsyn IG, Kartashov MY, Tyuten Kov OY, Protopopova EV, Romanenko VN, Chausov EV, Gashkov SI, et al. 2014. Surveillance of tick-borne encephalitis virus in wild birds and ticks in Tomsk City and its suburbs (Western Siberia). Ticks Tick Borne Dis. 5:145–151. [DOI] [PubMed] [Google Scholar]
- Nosek J. 1971. The ecology, bionomics and behaviour of Haemaphysalis (Haemaphysalis) concinna tick. Z Parasitenkd. 36:233–241. [DOI] [PubMed] [Google Scholar]
- Švehlová A, Berthová L, Sallay B, Boldiš V, Sparagano OAE, Špitalská E.. 2014. Sympatric occurrence of Ixodes ricinus, Dermacentor reticulatus and Haemaphysalis concinna ticks and Rickettsia and Babesia species in Slovakia. Ticks Tick-Borne Dis. 5:600–605. [DOI] [PubMed] [Google Scholar]
- Teng KF, Jiang ZJ, editors. 1991. Economic insect fauna of China, Fasc 39, Acari: Ixodidae. Beijing: Science Press; p. 359. [Google Scholar]
- Yamasaki H, Ohmae H, Kuramochi T.. 2012. Complete mitochondrial genomes of Diplogonoporus balaenopterae and Diplogonoporus grandis (Cestoda: Diphyllobothriidae) and clarification of their taxonomic relationships. Parasitol Int. 61:260–266. [DOI] [PubMed] [Google Scholar]