Abstract
In this study, the complete mitochondrial genome of human lung fluke, Paragonimus kellicotti, was recovered through Illumina sequencing data. This complete mitochondrial genome of P. kellicotti is 13,927 bp in length and has a base composition of A (16.6%), T (41.8%), C (13.%), and G (28.4%), demonstrating an obvious bias of high AT content (58.4%). The mitochondrial genome contains a typically conserved structure, encoding 12 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (12S rRNA and 16S rRNA), and a control region (D-loop region). All PCGs were located on the H-strand. ND4 gene and ND4L gene were overlapped by 39 bp. The nucleotide sequence of 12 PCGs of P. heterotremus and other 10 parasite species were used for phylogenetic analysis. The result indicated P. heterotremus a relative close relationship with species P. westermani (AF219379.2).
Keywords: Paragonimus kellicotti, mitochondril genome, assembly, phylogeny
Paragonimus kellicotti, the North American lung fluke, which is widely distributed in small carnivores such as mink, skunks, otters, and other mammals that feed on crayfish, is a species of trematode in the genus Paragonimus. Until now, the organelle genome information of P. kellicotti is still limited. In this study, the complete mitochondrial genome of P. kellicotti was recovered through Illumina sequencing data. This complete mitochondrial genome can be subsequently used for clinical diagnosis and provide valuable insight into phylogeny relationship among Paragonimus species.
The eggs of P. kellicotti was collected from crayfish found in Black River (37°26′34′′N, 90°50′48′′W) in the Ozark region of southeastern Missouri. Adult worms were obtained by feeding other crafish with metacercariae. Genomic DNA was extracted from adult worms using the commercial QiaAmp DNA extraction kit or DNeasy Tissue kit (Qiagen Inc.) according to the manufacturer’s instructions. The isolated DNA was stored at –20 °C in the functional lab of Institute for Translation Medicine in Qingdao University. The partial genomic DNA was then subjected to standard Hiseq 2500 library construction. A total of 15 Gb reads were obtained with average length of 150 bp. After quality filtration, the clean reads were assembled by SPAdes 3.9.1 (Bankevich et al. 2012) based on default settings.
We used another mitochondrial genome of P. westermani (AF219379.2) as a reference sequence to align the contigs and identify gaps. To fill the gap, Price (Ruby et al. 2013) and MITObim v1.8 (Hahn et al. 2013) were applied and Bandage (Wick et al. 2015) was used to identify the circular topology. The complete sequence was primarily annotated by ORF (Open Reading Frames) prediction in Unipro UGENE (Okonechnikov et al. 2012) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe et al. 1997). Other PCGs were verified by BLAST search on the NCBI website (http://blast.ncbi.nlm.nih.gov/), and manual correction for start and stop codons were conducted. The circular mitochondrial genome map was drawn using OrganellarGenomeDRAW (Lohse et al. 2007). This complete mitochondrial genome sequence together with gene annotations were submitted to GenBank under the accession numbers of MH322000.
The complete mitochondrial genome of P. kellicotti was 13,786 bp in length and has a base composition of A (16.7%), T (42.4%), C (12.6%), and G (28.2%), demonstrating an obvious bias of high AT content (59.1%). The mitochondrial genome contains a typically conserved structure, encoding 12 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (12S rRNA and 16S rRNA), and a control region (D-loop region). All PCGs were located on the H-strand. ND4 gene and ND4L gene were overlapped by 39 bp.
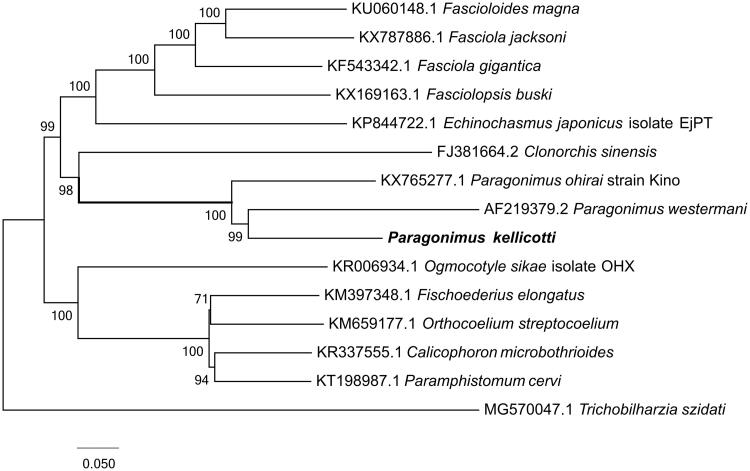
Phylogenetic analysis was constructed by applying 12 mitochondrial PCGs with other 14 closely related taxa. The whole genome alignment was constructed by HomBlocks (Bi et al. 2018) and verified by MAFFT (Katoh and Standley 2005). Finally, conserved regions were picked out by Gblocks 0.91b (Castresana 2002) to construct concatenated nucleotide sequences. Phylogenetic tree constructed using RAxML version 8.1.12 (Stamatakis 2014) was shown in Figure 1. The relationships among the 15 taxa were fully resolved with high bootstrap values among all nodes. Paragonimus kellicotti was clustered into the group of genus Paragonimus and exhibited a relative close genetic distance with P. westermani (AF219379.2).
Figure 1.
The ML tree of the 15 species from parasites was constructed based on 12 commmon proteincoding genes. Numbers beside divergence nodes are percents of 1000 bootstrap values. The analyzed species and corresponding Genebank accession numbers were also labbeled.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2002. Gblocks, v. 0.91 b [accessed 2012 February 6]. http://molevol.cmima.csic.es/castresana/Gblocks_server.html [Google Scholar]
- Hahn C, Lutz B, Bastien C. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- Ruby J, Graham P, Bellare JL. 2013. DeRisi PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 Genes Genomes Genetics 5:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]