Abstract
Rubber is a fascinating material in both industry and daily life. The development of elastomeric material in nanotechnology is imperative due to its economic and technological potential. By virtue of their distinctive physicochemical properties, nucleic acids have been extensively explored in material science. Phi29 DNA packaging motor contains a 3WJ with three angles of 97°, 125°, and 138°. Here, the rubber-like property of RNA architectures was investigated using optical tweezers and in vivo imaging technologies. The 3WJ 97° interior angle was contracted or stretched to 60°, 90°, and 108° at will to build elegant RNA triangles, squares, pentagons, cubes, tetrahedrons, dendrimers, and prisms. RNA nanoarchitectures was stretchable and shrinkable by optical tweezer with multiple extension and relaxation repeats like a rubber. Comparing to gold and iron nanoparticles with the same size, RNA nanoparticles display stronger cancer-targeting outcomes while less accumulation in healthy organs. Generally, the upper limit of renal excretion is 5.5-nm, however, the 5, 10, and 20-nm RNA nanoparticles passed the renal filtration and resume their original structure identified in urine. These findings solve two previous mysteries: 1) why RNA nanoparticles have unusual high tumor targeting efficiency since their rubber or amoeba-like deformation property enables them to squeeze out of the leaky vasculature to improve EPR effect; 2) why RNA nanoparticles remain nontoxic since they can be rapidly cleared from the body via renal excretion into urine with little accumulation in the body. Considering its controllable shape and size plus its rubber-like property, RNA holds great promises for industrial and biomedical applications especially in cancer therapeutics delivery.
Keywords: RNA nanotechnology, pRNA 3WJ motif, elastomer, RNA rubber, rubber-like material, optical tweezers
Graphical Abstract
RNA, DNA, and their chemical derivatives are important starting materials in nanotechnology,1 spanning a wide range of applications such as targeted drug delivery,2 nanorobots,3 nanocircuits,4 and many others.5–8 The programmable self-assembly of nucleic acids, along with their distinctive physical and chemical properties,9–16 have made these applications possible. For these applications, different modifications of natural bases are performed to improve nucleic acid characteristics such as chemical and thermodynamic stability.17 The 2’-F modification of RNA, where a fluorine is present at the 2’ ribose position,17 is commonly used to prevent enzymatic degradation by ubiquitous RNases, greatly increases the potential application of RNA in many settings.18,19 Elasticity,20–27 which is another important property of nucleic acid polymers,28–31 allows them to behave like a rubber with nanoscale control in dimension.32 As a widely used material in both industry and daily life, rubber has been extensively studied regarding its fabrication, mechanism, and rubbery property.33,34 The meaning of “Elastic” is similar to the implication of “Rubbery” to some extent. Elastic refers to the physical property (genotype), while rubbery is more or less a general term, as here referring to the biological effect (phenotype). This report deals with a biological system, and to make it easier to understand by both the cancer biologist and physicist, we interchange both the elasticity and rubbery property.
Due to the mammoth economic and technological importance of rubberlike materials,34 the investigation and search for elastomers has been of great interest to the burgeoning field of nanotechnology.35 The Lego-like property RNA and DNA,36,37 which allows the bottom-up assembly of nanomaterials in material science, has generated further interest in elucidating the rubbery property of RNA nanoarchitectures using optical tweezers.38,39 The typical feature of rubber is well dictated by its ability to extend under applied external force and then return to its original shape upon release of the external force.35 The structural flexibility of the RNA motif allows for the modular assembly of nanostructures with stretchable angles in different conformations.40
Naturally folded RNA molecules typically contain a large variety of single stranded loops, knots or cohesive ends for intra-and/or intermolecular interactions, and these loops or knots can serve as ‘dovetails’ to connect different building blocks of “RNA Legos”, thus circumventing the need for an additional dowel. Loops, knots, and motifs have allowed for the construction of more complicated 3D architectures,41–43 and 2’F-RNA-based dimers, tetramers,44 triangles, squares,40,45 tetrahedrons,46 pentagons,40,42 cube,11 prisms,47 pyramids,48 dendrimers,49 micelles18,50 have all been reported.
It is well established that chemical and physical properties of nanoparticles play an important role in their In vitro and In vivo interaction, thus contributing to pharmacokinetics and biodistribution properties.51–53 The role of particle rubbery property on key functions including blood circulation time, biodistribution, antibody-mediated targeting, endocytosis, and phagocytosis has been well demonstrated.54,55 The application of various RNA nanostructures as a carrier in cancer therapy has been extensively studied. We’ve demonstrated that RNA nanoparticles based on 3WJ motif of high thermodynamic stability, controllable size, shape, and stoichiometry possess favorable tumor inhibition efficiency and limited toxicity and immunogenicity.56–58 However, the impact of the rubbery property of RNA nanostructures needs to be systematically studied. Here we have demonstrated the stretching ability and rubbery property of RNA architectures and exponential Force-Extension curve similar to rubber under external force. The rubbery property of RNA nanoparticles is demonstrated In vivo by comparing the retention time in the tumor, kidney, and liver. The rubbery behavior of RNA nanoparticles was further supported by the urinary excretion of RNA 4WJ larger than glomerular filtration capacity.59
Results and Discussions
Demonstration of RNA as elastomer by reshaping the 97° angle of the pRNA 3WJ into 60°, 90°, and 108° to build various RNA hierarchical architecture
The crystal structure of the pRNA 3WJ of bacteriophage phi29 DNA packaging motor reveals a planar shape. Extension of each of the three double stranded arms of the 3WJ revealed three angles of 97°, 125°, and 138°, respectively (Fig. 1). The 97° angle of the 3WJ can be contracted or stretched to 60°, 90°, and 108° at will in silico design to build elegant RNA triangles, squares, pentagons, cubes, tetrahedrons, dendrimers, and prisms, demonstrating that RNA can serve as elastomer in Lego construction.36,40,46–48,60–62 Each RNA nanostructure contained a pRNA 3WJ motif at each vertex. Interestingly, after being incorporated into these RNA 2D or 3D nanostructures, the three strands of the 3WJ adopt different angles to accommodate the structure, supporting the proposal that RNA displays rubbery property (Fig. 1).
Figure 1. Construction and characterization of different RNA nanoparticles based on 3WJ.
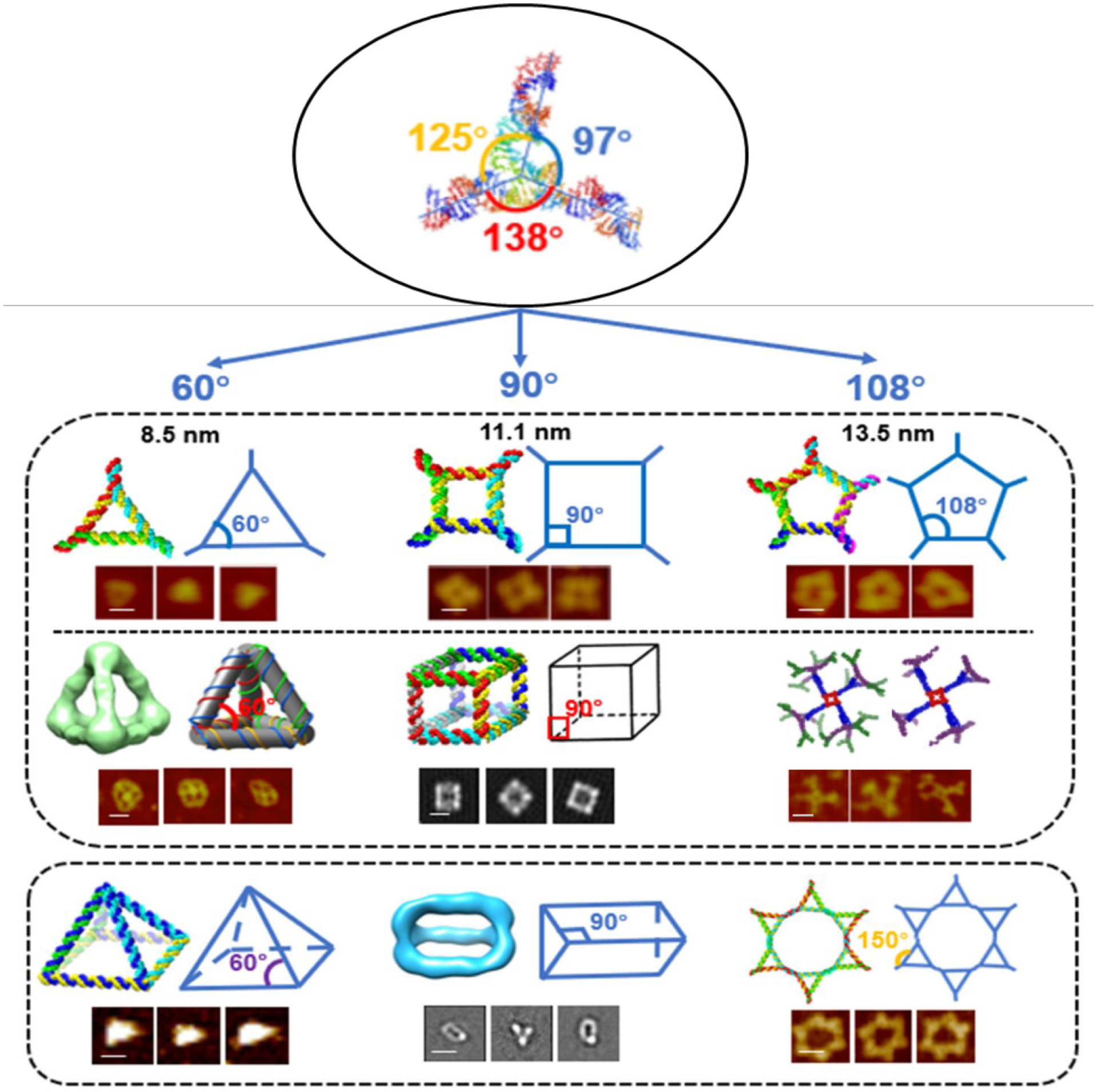
Angles of original 3WJ were stretched to accommodate different shapes of 2D or 3D RNA nanoparticles. In each of the structure, schematic and AFM/Cryo-EM images are shown. Figures are adapted with permission from previous publications.40,46–48,62 Scale bar is 10 nm in each image.
Exponential Force-Extension Curve Similar to Rubber Under External Force
Several physical mechanisms are proposed here that elucidate the rubbery force within the chains of the square when RNA or DNA nanostructures are being stretched. One proposed mechanism is the change in entropy, and the second is related to the distortion of the molecular bond angles along the backbone chain.63,64 These mechanisms have been observed when the DNA or RNA was stretched under force. Initially, the polymer is quite flexible and thus the force could increase at a lower rate with respect to the strain. At an intermediate process, the requirement for force should be higher to cause the same extend of stretching. Later, as the polymer approaches the breaking point, its stiffness increases, thus requiring much higher force to stretch the polymer. If the force is released before the breaking point, the molecule can be restored to its original shape. Comparison of the stress/strain curve of many other elastomers from literature65–67 and the force/extension curve of the dsDNA construct (Fig. 2) revealed similar slopes and shapes, leading to the extrapolation that nucleic acid polymers are potentially elastomers.
Fig. 2. Repeated stretching-relaxing of nucleic acid polymer.
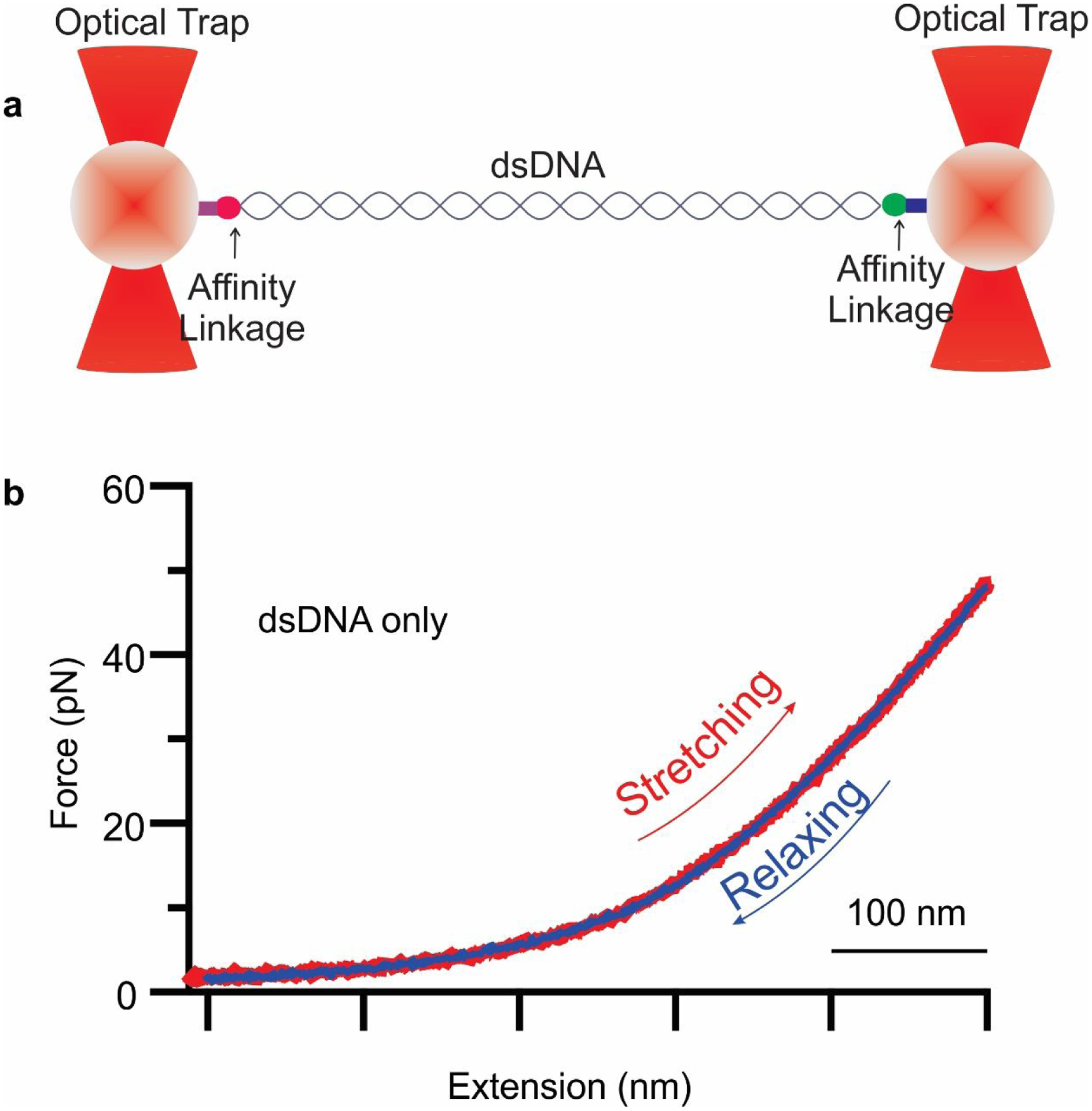
a. Schematic diagram of dual trap optical tweezers with a tethered dsDNA construct b. Force-extension curve of dsDNA construct. Stretching and relaxing of the same molecule repeatedly showing rubber-like property of nucleic acids polymer.
Rubber is known for its rubbery property: i.e., the ability to be stretched to an extended length under an external force and revert to its original shape when the force is released.68 Stretching repeatedly to relatively great lengths and then relaxing to restore to the original position is a common property of rubbery materials.69,70 Here we placed a 5 kb dsDNA construct between two polystyrene beads via affinity linkage of biotin-streptavidin and digoxygenin-antidigoxigenin on opposite ends. One bead was fixed, and another bead was moved by a steerable mirror to apply external force on the single molecule (Fig. 2a). This resulted in a force-extension exponential curve with stretching and relaxing. Stretching and relaxing of a single molecule were performed repeatedly many times before reaching the melting plateau of approximately 65 pN (Fig. 2b). This repeated stretching to longer distances, followed by relaxing back to the original position suggests the rubbery property of nucleic acid polymers.
RNA Rubbery Property Investigated by Optical Tweezers
The rubbery property of RNA material was demonstrated by force and extension measurements with dual beam optical tweezers. RNA squares were constructed using previously reported methods.40 Specifically, five strands were mixed at a stoichiometric ratio to self-assemble. By incorporation of phi29 3WJ sequences at each corner, the 2D nanosquare was assembled with high thermodynamic stability. Two overhangs were created diagonally for dsDNA handle ligation, facilitating the optical tweezer measurement. The square RNA is sandwiched between two dsDNA handles by ligation to attach to the polystyrene beads at the ends through a biotin-streptavidin and dig-antidigoxigenin linkage separately (Fig. 3a). Force ramping experiments were performed along the vertices of each square, using a loading rate of 5.5 pN/s to gradually increase the force by moving one of the trapped particles away from the other using a steerable mirror (Fig. 3a). A change in conformation of the square was demonstrated by a sudden jump in force-extension curves recorded during the experiments (Fig. 3b). The RNA conformational changes resulting from the applied force reverted to the original conformation slowly as shown in the relaxing curve (Fig. 3b). This reversible feature of the conformational change of the RNA nano-square demonstrates the rubbery property of RNA nanoarchitectures.
Fig. 3. Rubbery property of RNA and DNA nanosquare by optical tweezer analysis.
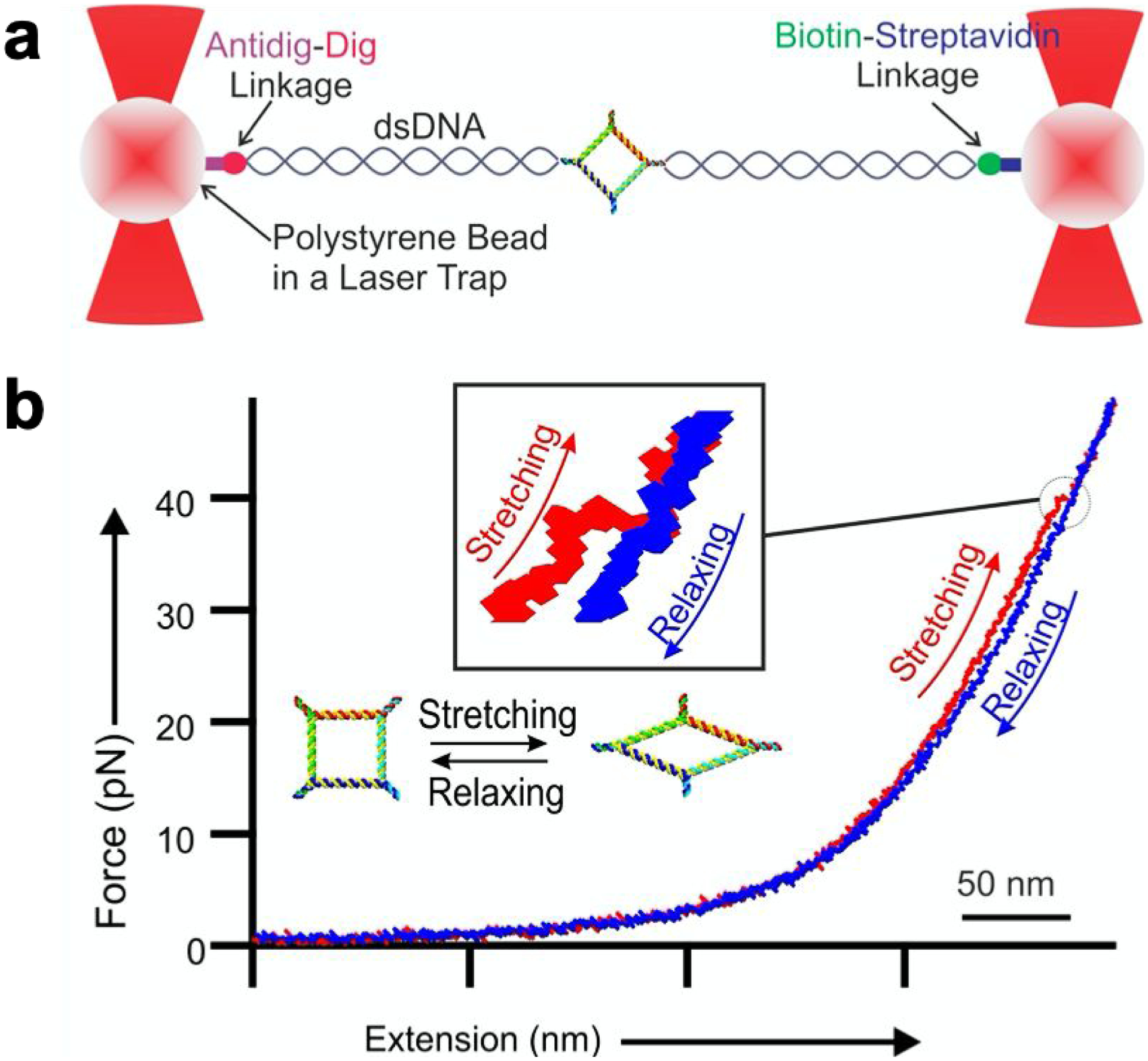
a. Schematic diagram of dual trap optical tweezers with a tethered nanosquare construct of DNA or RNA sandwiched between two dsDNA handles via affinity linkers. b. A typical force-extension curve for stretching (red) and relaxing (blue) of square nanoparticle. Inset shows magnified view of conformational change of the square nanoparticle. The 50 nm scale bar is for the x-axis of force extension curve.
From the force-extension curve, a change in extension was quantified to assess the stretchiness of the particles. The change in extension at a given force was calculated as a difference between the extension of stretching and relaxing curves. (Fig. 4a). It was found that among the RNA and DNA squares, the change in extension for RNA is longer at 4.1 ± 0.10 nm, compared to 2.99 ± 0.03 nm for DNA. It is well known that RNA and DNA exhibit differences in their elasticity due to their respective A- and B- form helical structures. Their Young’s moduli are well-studied by determining the extension and releasing force measured by force spectroscopy.31 This difference could be due to the natural difference of RNA and DNA. The A form and B form of RNA are different concerning the structure of helix and a different number of nucleotides per helical turn of RNA and DNA as reported previously.31,72–73 This difference specifically may have been contributed by the C3’-endo and C2’-endo conformation of the A-form and B-form RNA and DNA, respectively. The C3’-end form has phosphate to phosphate distance of 0.59 nm while that for the C2’-endo form on DNA is 0.7 nm. The compact structure of the RNA may have contributed to the longer change in extension for the nanosquare made of dsRNA. This result demonstrates the highly flexible and more rubbery behavior of the RNA nanoarchitecture compared to its DNA counterpart.
Fig. 4. Histograms of change in extension and force measurement for RNA (Top), and DNA (bottom).
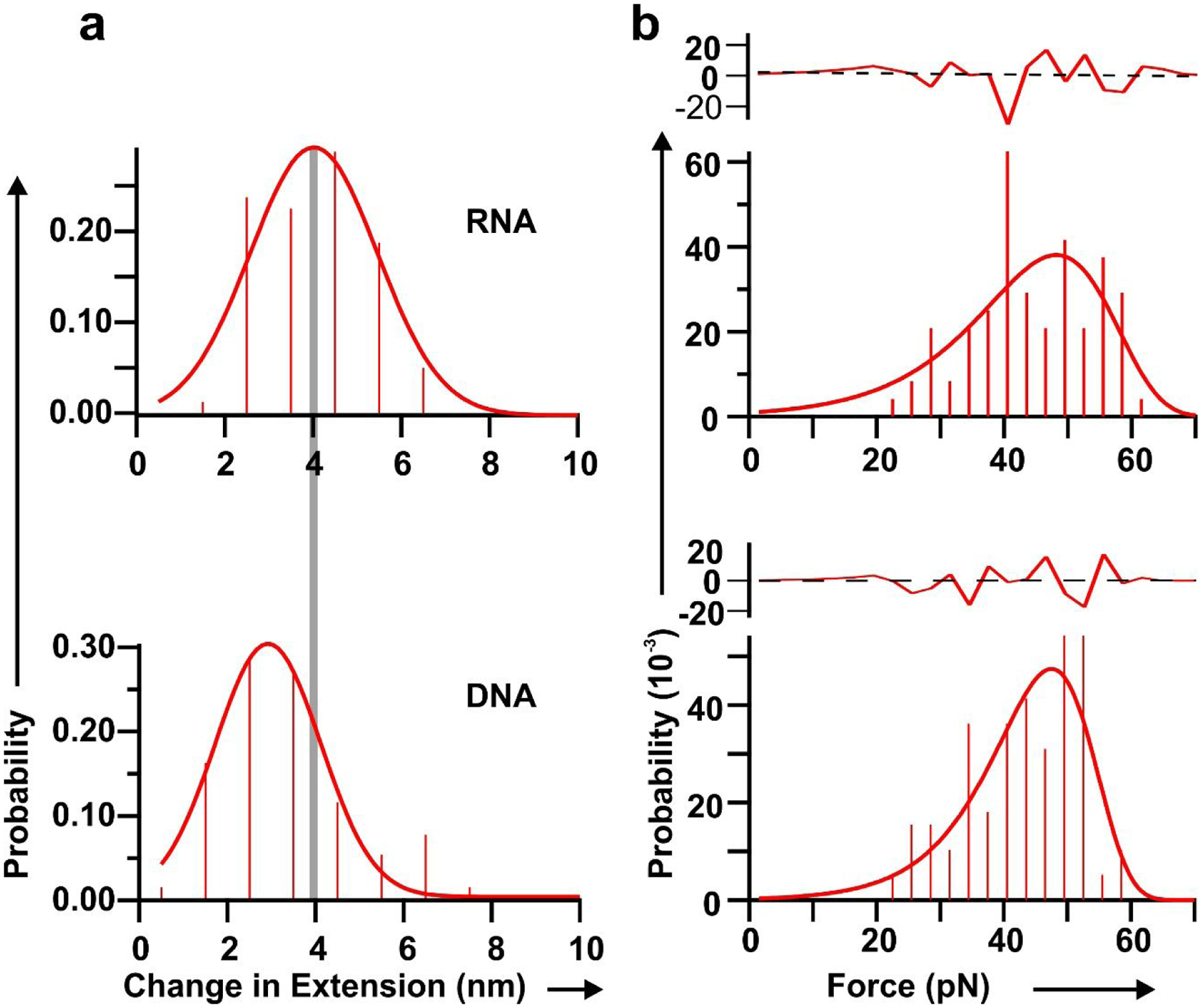
a. Change in extension from change in conformation. The grey line depicts the center of the histogram peaks. b. Histograms of force to change conformation measured from force-extension curves for different nano-squares. Each force histogram is fitted with the Dudko equation71 (solid line). Residuals of fitting are shown above each histogram. The black dotted lines depict average values.
A plot of force obtained from the force-extension curve corresponding to the change in the conformation of the nano-square depicts the mechanical stability of the particle. The plot of force shows high mechanical stability of the RNA, and DNA squares, at nearly 45 pN (Fig. 4b). Having a similar force profile but a different change in extension demonstrates the more stretchable property of RNA nanosquare under similar stress. Kinetics of change in conformation was analyzed by fitting the force histograms of each square to an equation proposed by Dudko et al.74 The solid red line shown in Fig. 4 represents the fitting. Results show that the change in activation energy is smallest for RNA as shown in Table 1. RNA also has a faster change in conformation, as demonstrated by its conformation change rate constant, and the smallest distance to the transition state from the state prior to the change in conformation. The lower activation energy required to change the conformation of RNA may explain the more flexible nature of the RNA nano-squares under tension.
Table 1 |.
Conformation change force (Force, pN), change in extension (ΔX, nm), conformation change rate constant (kcc, per sec), distance to the transition state (x†, nm) and conformation change energy barrier (ΔG†, kcal per mol), for DNA and RNA squares.
| FORCE | ΔX | Kcc | X† | ΔG† | |
|---|---|---|---|---|---|
| DNA | 44.2±2.3 | 2.99±0.03 | 0.0016±0.0007 | 0.132±0.008 | 13.5±0.5 |
| RNA | 44.6±4.1 | 4.1±0.10 | 0.005±0.0030 | 0.10±0.020 | 11.4±1.4 |
Demonstration of the rubbery property of RNA nanoparticles by comparing the retention time in tumor, kidney, and liver
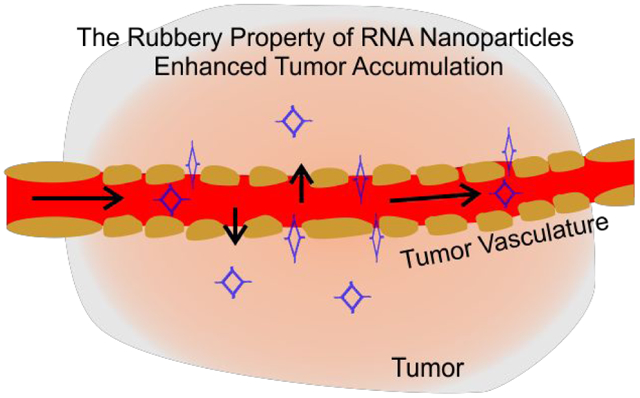
It has been widely studied that nanoparticles display EPR (enhanced permeability and retention) effect due to the nano-scale size. If the strong EPR effect and strong tumor accumulation are due to the special property of RNA rubbery property as proposed in this report, then the EPR effect of iron or gold nanoparticles should be lower than the RNA nanoparticles of same size.
The EPR effect has been demonstrated as the major pathway for nano-scale particles accumulation at the tumor site.75,76 The rapid growth of a tumor results in poor production of blood vessels that are incomplete and leaky77 Recent studies suggested that at least some human cancers contain leaking blood vessels,77 providing holes tens of nm in size for nanoparticle penetration.78 These characteristics account for favorable penetration and retention of nanoparticles in tumors.
We have previously demonstrated that, based on high thermodynamic stability, controllable size, shape, and stoichiometry, RNA nanoparticles can serve as therapeutics delivery vehicles harboring chemical drugs and RNA therapeutic agents for tumor targeting even without the tumor binding ligands.57 The strong EPR effect of RNA nanoparticles has been extensively exploited as a drug delivery platform.79–81 The size of RNA nanoparticles results in a strong EPR effect while avoiding entrapment in the liver and spleen by the mononuclear phagocytic system (MPS) recognition.82–84 Systemically injected RNA nanoparticles strongly and specifically bind to cancers with reduced accumulation in the liver, lung, or any other vital organs or tissues.80,85–87
It is expected that the phenomenon of tumor accumulation without the use of targeting ligands is due to EPR effect which leads to accumulation of RNA nanoparticles in the cancer vasculature without cell entry.50,88 To further investigate the mechanism for such favorable accumulation in the cancer vasculature, we quantitatively compare tumor uptake, organ retention, and kidney clearance of RNA nanoparticles to solid inorganic nanostructures, such as iron, and gold of similar or same size (Fig. 5). To compare the EPR effect and the proposed rubbery property of RNA nanoparticles, NIR fluorescent dye labeled a 6 nm RNA 3WJs, 10 nm 4WJs, 12 nm 6WJs (Fig. S1), 10 nm gold nanoparticles, and 10 nm iron nanoparticles were injected intravenously into mice. The In vivo organ distributions of various RNA nanoparticles and their inorganic nanoparticles counterpart were examined 8 hours post injection. The fluorescent intensity of Ex vivo entire organ as well as homogenized organ sample were quantified. It was found that at 8 hours, RNA nanoparticles of different stoichiometry and the iron or gold nanoparticles exhibit big differences in organ and tumor retention (Fig. 5a). After normalizing the radiation efficiency by organ weight, 4WJ-RNA nanoparticles possess a higher tumor to liver and tumor to kidney ratio (Fig. 5b). The homogenized organ sample data is consistent with the Ex vivo result (Fig. 5c), which further demonstrates the favorable biodistribution profile of RNA nanostructures. Interestingly, the tumor to organ fluorescence ratio, of the 10 nm RNA nanoparticles was much higher than the iron and gold with the same size of 10 nm. These results support the hypothesis that RNA nanoparticles display rubbery property, leading to the stronger EPR effect and enhanced vessel extravasation in tumor targeting.
Fig. 5. Demonstration of the rubbery property of RNA nanoparticles by comparing the retention time in tumor, kidney and liver.
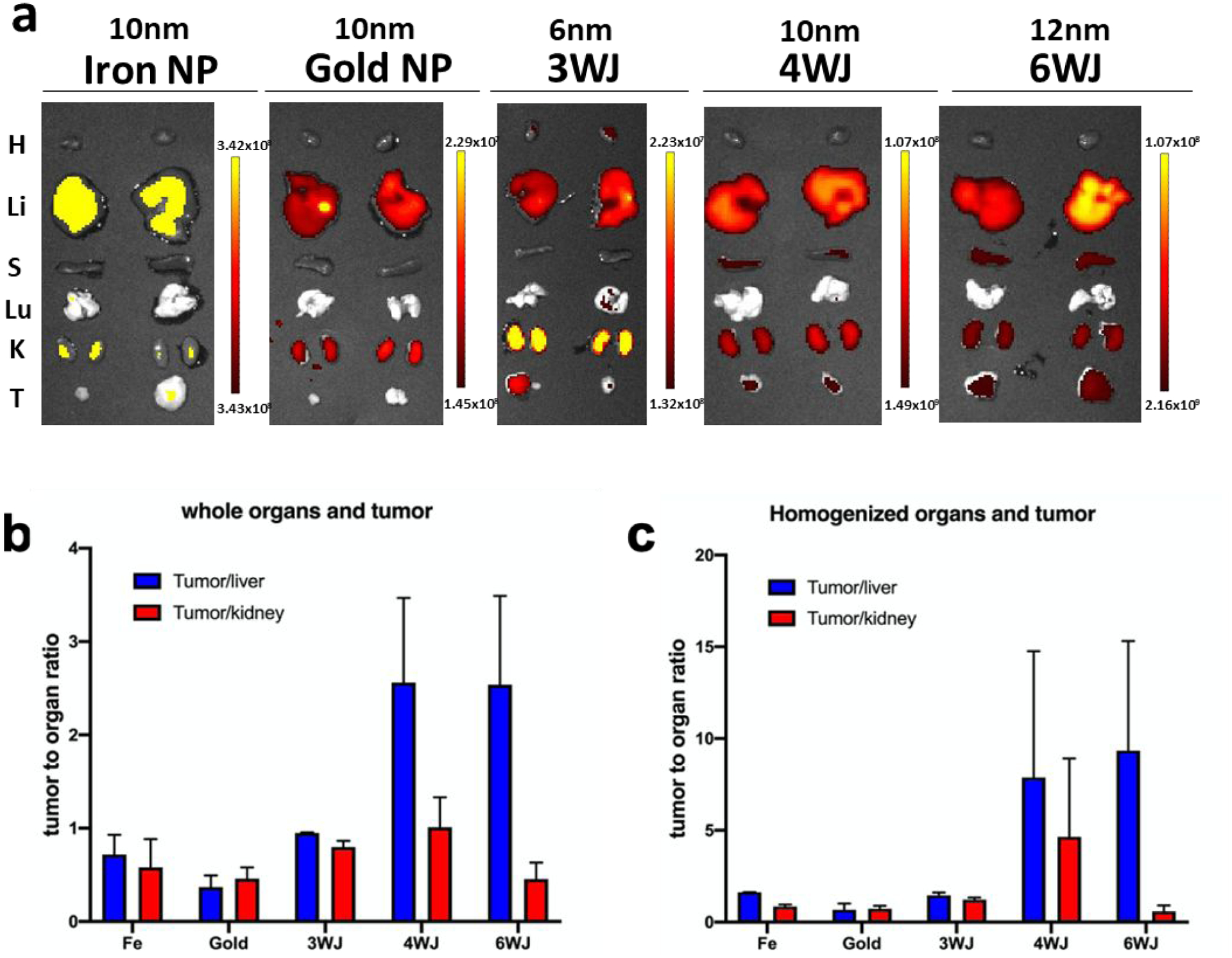
a. The Cy5.5 labeled nanoparticles were detected by Ex vivo organ 8 hr post-injection in mice bearing KB xenograft (T: tumor, H: heart, S: spleen, Lu: lung, K: kidney, and Li: liver; Color scale: radiant efficiency, [p s−1 cm−2 sr−1] / [μW cm−2]). b. Quantitative analysis of whole body biodistribution to quantify the ratio of tumor to liver and tumor to kidney using images from a. c. Quantitative analysis of biodistribution in tumor to liver and tumor to kidney ratio, quantified from the homogenized organ sample.
Demonstration of the Rubbery Property of RNA Nanoparticles by Whole Body Imaging for Body Clearance
Chemical or nanoparticle excretion implies the clearance of the intact complex from the body. Nonvolatile chemicals or nanoparticles are excreted mainly by renal excretion, a process in which these small complexes are filtrated via the kidney into the bladder and ultimately into the urine. In the whole-body imaging study, the nanoparticles of different sizes were injected intravenously into the mice, and the distribution and clearance of the nanoparticles were monitored by whole body imaging. Mice were imaged for whole body fluorescence at time points of 0.5, 1, 2, 4, 8, and 12 hrs (Fig. 6). The larger the nanoparticle size, the longer the circulation time was observed with final clearance by renal excretion from the body. Even the 20 nm squares were removed from renal system which has filtration cut off < 5.5 nm. These results suggested the flexible nature of the nanoparticles may allowing them to pass through the kidney due to the rubbery property of RNA nanoparticles.
Fig. 6. Demonstration of the rubbery property of RNA nanoparticles by whole body imaging for body clearance, the indirect evidence of renal excretion.
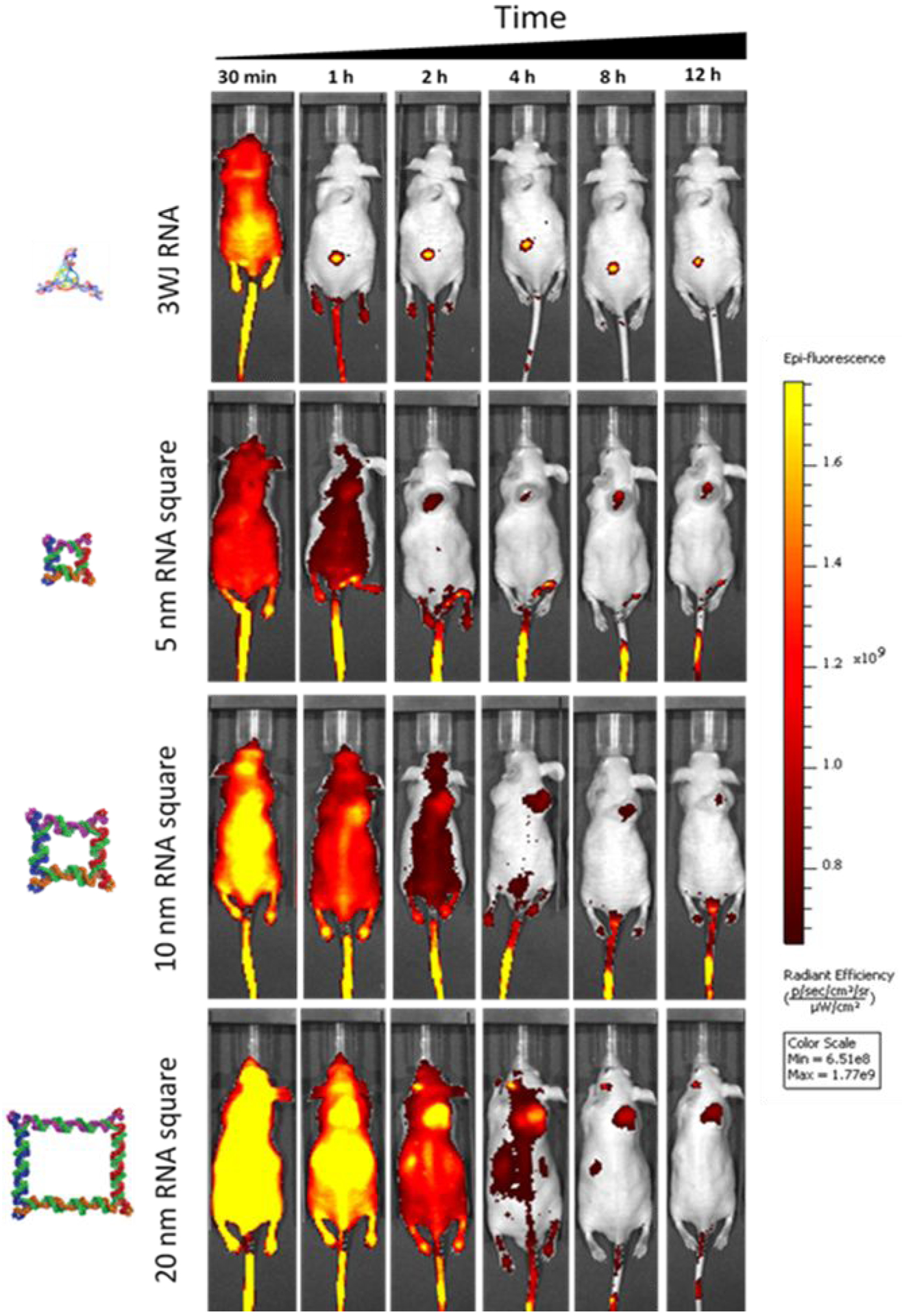
Longer circulation time with increasing nanoparticles size.
Demonstration of The Rubbery Property of RNA Nanoparticles Through Renal Excretion by Comparing Tumor and Kidney Retention Time
In our earlier research, we found that the biodistribution of RNA nanoparticles is size and shape dependent.89 We also reported the timely clearance of the RNA nanoparticles in organs (Fig. 4 in Jasinski. D, 2018).89 In our current investigation, we looked at this set of data with another view and interpretation: comparing the retention and clearance time in the tumor and kidney. As reported earlier,89 mice were sacrificed and organ samples were collected and imaged for Ex vivo organ fluorescence at time points of 12 hours (left) and 24 hours (right), respectively (Fig. 7). Ex vivo organ images of 10 and 20 nm RNA squares suggested that the RNA nanoparticles larger than the 5.5 nm renal execration limit passed the kidney filtration. As evidenced, near-Infrared AF647 labeled 10 nm and 20 nm square were detected both in kidney and tumor 12 and 24 hours post injection; however, the kidney signal significantly reduced after 12 additional hours whereas the fluorescence in tumor had a minor reduction. This result suggests that these large RNA squares were excreted through kidney filtration since they stayed in the tumor with longer retention time in comparison to that in the kidney. Since the upper limit of the renal excretion is about 5.5 nm, the renal excretion and without body accumulation of the nonvolatile and non-degradable RNA nanoparticles could hind their rubbery property.
Fig. 7. Demonstration of the rubbery property of RNA nanoparticles through renal excretion by comparing tumor and kidney retention time using Ex vivo organ images.
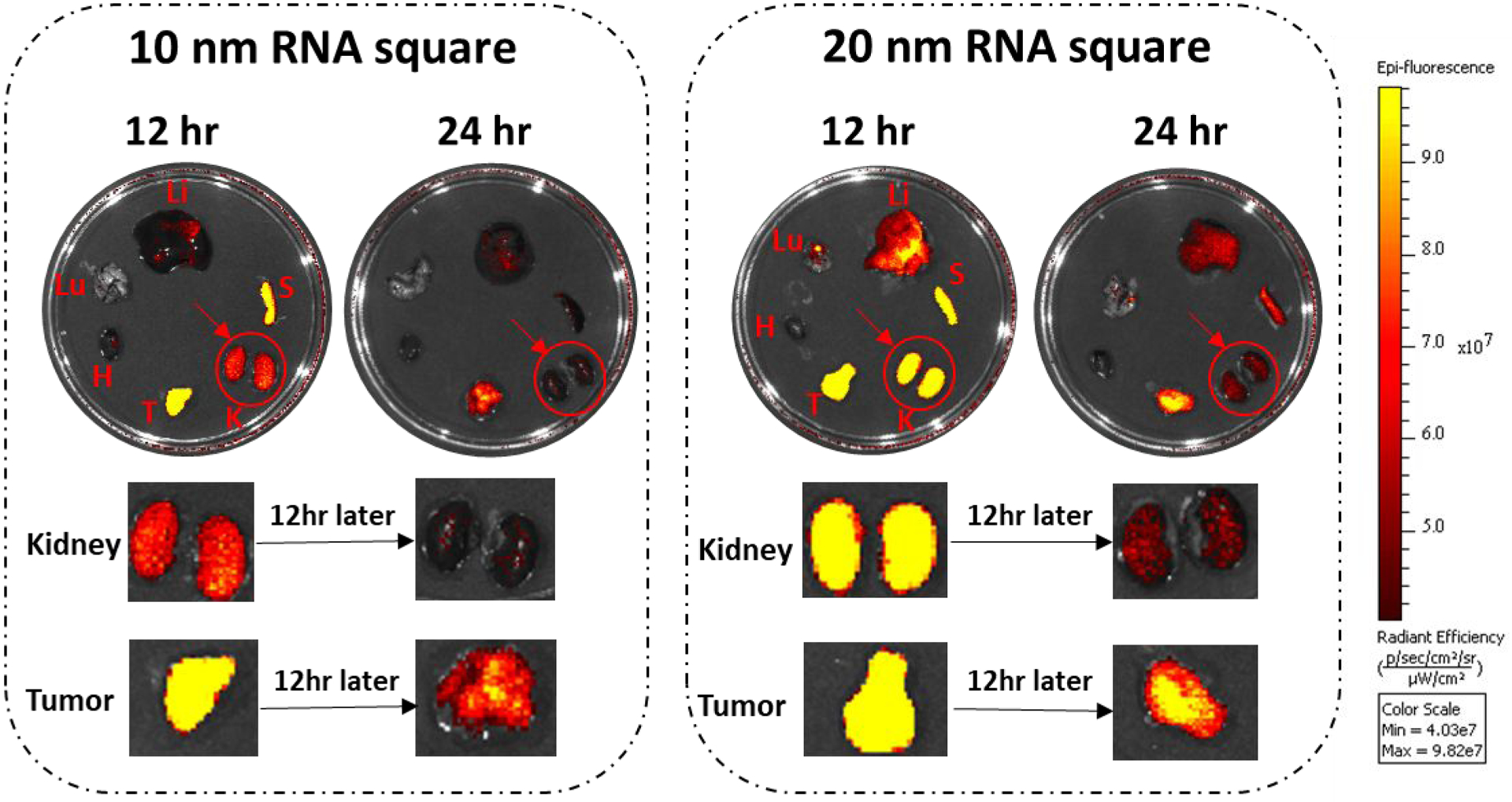
The near-infrared AF647 labelled 10 nm and 20 nm squares were detected in the kidney 12 and 24 hrs post systemic injection. Both the 10 and 20 nm RNA squares were excreted through kidney filtration, while with longer retention time in tumor by comparing the organ intensity after 12 hrs.
Demonstration of The Rubbery Property of RNA Nanoparticles by Detecting the RNA Nanoparticles in Urine appeared as their original size before injection
As aforementioned, renal excretion is a process in which chemicals or nanoparticles are subjected to kidney glomerular filtration into the bladder and ultimately into the urine. The upper limit of renal excretion is 5.5 nm. Although the data from the whole-body and organ imaging suggests the rubbery property of RNA nanoparticles based on the presumption that renal excretion is the major pathway of RNA nanoparticles eliminated from the body. However, the clearance of the fluorescent signals from the body cannot completely exclude the possibility that RNA nanoparticles have been degraded or metabolized in the body. Thus, the clearance and lack of or reduced organ accumulation as assessed by whole body imaging cannot fully support the rubbery property hypothesis.
To further confirm the rubbery property of RNA nanoparticles during renal excretion, we injected the nanoparticles of different sizes and collected mice urine samples for further investigation. In our study, double strand(ds-), 3WJ- and 4WJ- RNA nanoparticles were injected into mice. Urine was collected 0.5 hr post systematic injection and assayed by 12% polyacrylamide TBE (89 mM Tris base, 200 mM boric acid, and 2 mM EDTA) gel electrophoresis. Intact 5, 6, and 10 nm RNA nanoparticles were detected in the gel from urine samples (Fig. 8), suggesting direct kidney filtration of RNA nanoparticles larger than the kidney filtration limit.
Fig. 8. Demonstration of the rubbery property of dsRNA and RNA nanoparticles by renal excretion.
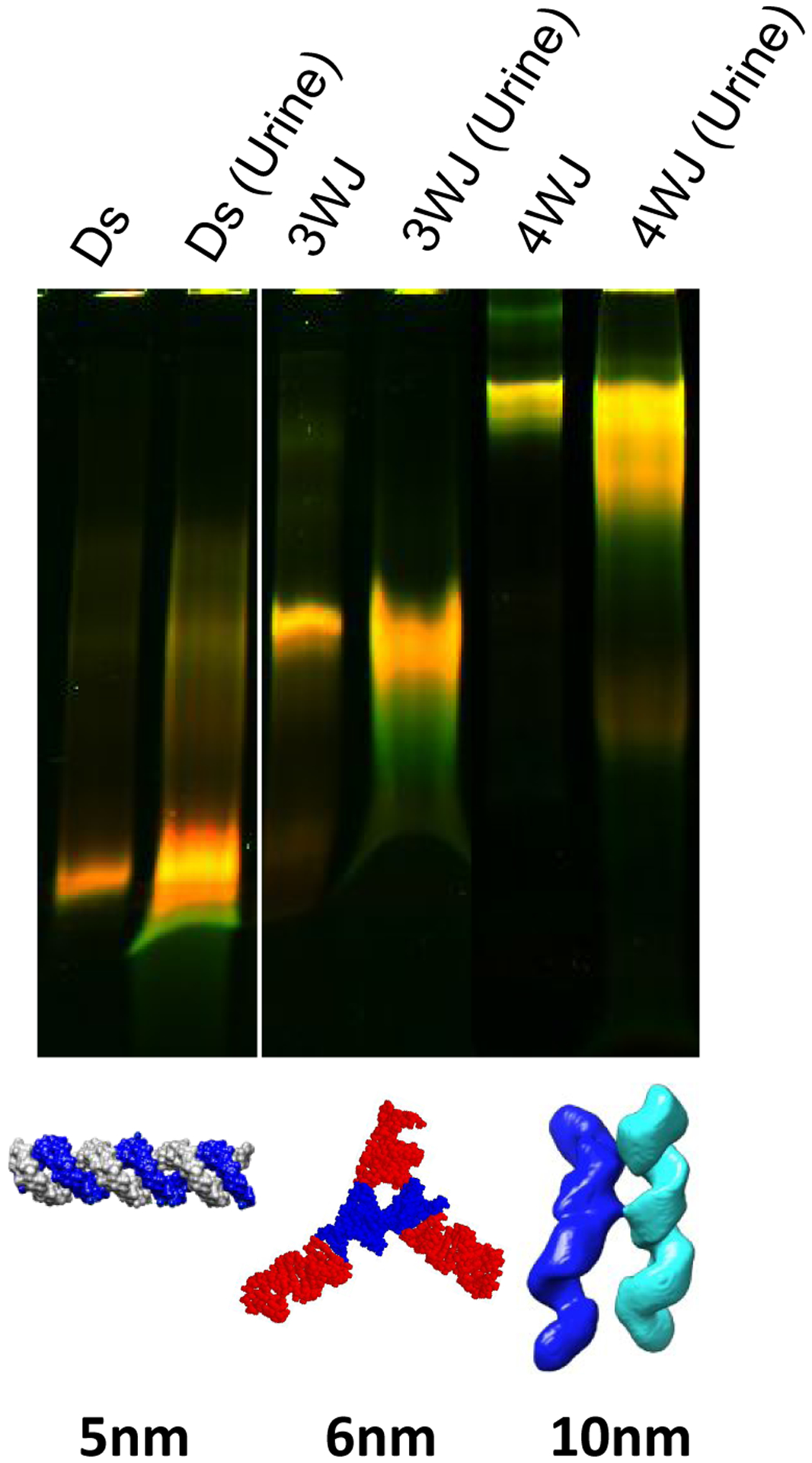
Near-infrared AF647 labelled dsRNA, RNA 3WJ and 4WJ were found in mice urine assayed by 12% native gel 0.5 hr post IV injection.
Direct glomerular filtration is highly dependent on molecule size and structure. It is reported that the physiologic pore size is 4.5–5 nm after considering the combined effects of each layer of the glomerular capillary wall. Smaller molecules with a hydrodynamic diameter (HD) < 5.5 nm are filtered directly, while larger molecules cannot be eliminated by the glomerulus.59,90–92 The 3WJ and 4WJ RNA nanoparticles are larger than the filtration slit. However, they were excreted very quickly by direct kidney filtration as the complete nanoparticles shown in the gel (Fig. 8), suggesting that the RNA nanoparticles may tune their shape to fit and go through the filtration slit and recover back to its original structure like rubber after passing through the pore.
Conclusion
The angle between RNA strands can be contracted or stretched to different degrees to build RNA triangles, squares, pentagons, cubes. tetrahedrons, dendrimers, and prisms, demonstrating that RNA can serve as an elastomer. RNA squares are stretchable and shrinkable as shown with multiple extension and relaxation repeats with optical tweezers, demonstrating the rubbery property. The results from the comparison of the retention time in tumor, kidney, and liver suggested the greater EPR effect of RNA nanoparticles and reduced vital organ accumulation. The results from the comparison of tumor and kidney retention time as well as excretion of various sized particles beyond the renal filtration limit suggested that RNA structure can change their conformation during renal filtration and resume to the original structure observed in the urine. Urinary excretion of RNA 4WJ with size larger than the pore size of glomerular filtration capacity demonstrates the rubbery nature of RNA nanoparticles. The findings solve the previous puzzle of why RNA nanoparticles have an unusually high retention rate in a tumor. This is due to the amoeba-like deformation property of RNA nanoparticles in order to squeeze through leaky vessels of the tumor vasculature improving the EPR effect. Additionally, the amoeba-like property allows the intact RNA nanoparticles to squeeze through renal filters for urine excretion resulting in no toxicity. Considering its controllable shape and size as well as rubbery property, RNA nanoarchitecture holds great promises as elastomer for industrial and biomedical applications especially in nanorobotics as well as for a drug cargo carrier for targeted delivery.
Materials and Methods
Materials
All chemicals were purchased from either Fisher Scientific or Sigma with purities >99.0%. DNA oligomers were purchased from Integrated DNA Technologies (www.idtdna.com) and purified with denaturing polyacrylamide gel electrophoresis (PAGE). RNA was synthesized in the lab. Unless specified otherwise, all enzymes were obtained from New England Biolabs (www.neb.com). Streptavidin coated and anti-digoxigenin coated polystyrene beads were purchased from Spherotech (Lake Forest, IL, USA).
Preparation of the RNA nanoparticles
DNA, and RNA squares were prepared as described previously89 (Table S1 for sequence). DNA and RNA nanosquares for tweezers (Table S2 for sequence) prepared were sandwiched between two dsDNA strands of 2500 bp in length for mechanical experiments. Lambda DNA was used to prepare these two 2500-bp dsDNA handles by two separate PCRs using two pairs of primers. In one of the primers, biotin was introduced at the 5’ end. One of the handles was cut with BamHI and the other was cut with KpnI to make sticky ends for ligation with the square overhangs. These restriction enzyme sites were introduced during PCR using primers. In another handle, 3’ ends were labeled with digoxigenin using terminal transferase after PCR. The two 2500-bp dsDNA handles were mixed with the square particles in the equimolar ratio for ligation separately for DNA and RNA. These constructs were stored at −80 °C for future use without further purification.
RNA oligomers were chemically synthesized using typical phosphoramidite chemical synthesis on an automated oligo synthesizer. Each RNA strand was synthesized using 2’- fluorinated cytidine and uracil. 3WJ‐c strand was 5’ modified with a primary amine (Cat. No: 10 ‐ 1947 ‐ 90, Glen Research). Cyanine5.5 ‐ NHS ester were purchased from Lumiprobe. Conjugation reactions were carried out by mixing a 1:10 molar ratio of primary amine labeled 3WJ‐c: NHS Ester‐Fluorophore in 0.1 M sodium bicarbonate buffer, pH = 8.5. The conjugation reactions were incubated at room temperature for 16 hours while protected from light. Following incubation, the reactions were ethanol precipitated and washed twice with cold 75% ethanol to remove the majority of the unreacted fluorophore, facilitating purification. Sequences for all nanoparticles used in this study can be found in the Supplemental Information.
Single Molecule Force-Extension Measurement
Single molecule force ramping experiments were performed in home-built dual–trap optical tweezers93–95 having a 1064 nm laser at 23 °C in a 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 10 mM MgCl2. Streptavidin coated polystyrene beads preincubated with the square construct and the anti-digoxigenin coated polystyrene beads were separately trapped in two laser foci of the optical tweezers. The sample was tethered between the two beads through affinity interactions between digoxigenin-antibody and biotin-streptavidin complexes. When one bead was moved apart by a steerable mirror, the tension in the tether increased, which was recorded in the force-extension (F-X) curves using the Labview program (National Instruments, Austin, TX). We recorded the force range from 0–65 pN at 1000 Hz with a loading rate of 5.5 pN/s. The Single molecular nature of the DNA was confirmed by the observation of the 65 pN plateau in the F-X curve. The F-X curves were filtered using the Savitzky-Golay function with a time constant of 10 ms in a MATLAB program (The MathWorks, Natick, MA). Change in extension (Δx) at a given force was calculated from the difference between stretching and relaxing curves at that force. Wherever appropriate, three sets of experiments were performed to obtain standard deviations, which were reported in the main text or as error bars in Table 1.
The Dudko model, as shown in equation 1, was used to determine the transition energy barrier, transition distance, and rate constants for conformation change from the conformation change force histograms.
| (1) |
Where
Here k(F) and kcc are the conformation change rate constants at force F and 0 pN, respectively, x† is the transition state distance from the unstretched state, ΔG† is the height of the energy barrier and ν is the parameter to characterize the shape of the energy barrier (ν = 1/2 for a sharp, cusp-like barrier and ν = 2/3 for a softer, cubic barrier). Here we used both values and reported the average. Uncertainties in fitting values were estimated by separating data into three random groups.
In vivo Biodistribution Experiments
KB cells were cultured In vitro and subcutaneously injected under the skin of 4-week old male nude mice (Taconic). A total of 2 × 106 cells was injected into each mouse. Tumors were grown for 2 weeks before mice were injected IV through the tail vein. Mice were administered with PBS as a blank control and 100 uL of 15 uM nanoarchitecture. Mice were imaged for whole body fluorescence at time points of 0.5, 1, 2, 4, 8, 12, and 24 hrs with an In vivo Imaging System (IVIS) imager (Caliper Life Sciences, Waltham, MA). For this study, two mice were used for the 0.5 through 12 hr time points. At the 12 hrs time point, one mouse was sacrificed, and its organs extracted. At 24 hr the second mouse was sacrificed, and its organs extracted and imaged. Tumors, hearts, kidneys, livers, spleen, and lungs were extracted and imaged on the IVIS system.
Ex vivo Biodistribution Study using In vivo Imaging System (IVIS)
Male Nu/Nu mice with KB tumor xenograft were injected intravenously (IV) through tail vein injection. For this study, two mice were used for each nanoparticle. Mice were administered PBS as a blank control. 100 uL of 20 uM RNA nanoparticle samples or 1.5 mg/kg inorganic samples was injected. It is important to note that the concentrations of dyes were kept consistent in all In vivo experiments. At 8 hours mice were sacrificed and their hearts, kidneys, livers, spleen, and lungs collected and imaged for Cy5.5 fluorescent signal using an In vivo Imaging System (IVIS) imager (Caliper Life Sciences). The fluorescence imaging data of average radiant efficiency ([p s−1 cm−2 sr−1] [μW cm−2]−1) were quantitative by IVIS system program. All animal experiments were housed and performed in accordance with the Subcommittee on Research Animal Care of the Ohio State University guidelines approved by the Institutional Review Board.
Urine Excretion Profile
Ds, 3WJ and 4WJ nanoparticles were self-assembled in PBS buffer. 10uM 100uL nanoparticles in PBS buffer were prepared and the size of nanoparticles is measured by Dynamic Light Scattering (DLS) at 25 °C.
To study the urine excretion profile of RNA nanoparticles, 50 uM 100 uL AF647 labeled ds-, 3WJ- and 4WJ nanoparticles were injected into Balb/c mice by tail vein injection. Mice were housed on hydrophobic sands and urine sample would stay on the surface of the sand. The urine of mice was collected using pipette within 0.5 hr post injection. The samples were loaded to 12% Native Gel for electrophoresis under 150 V for 100 min in TBE buffer. The gel was visualized by Typhoon FLA 7000 (GE healthcare).
Supplementary Material
Acknowledgements
The research in P.G.’s lab was mainly supported by NCI/NIH grant U01CA207946, and partially support by NIBIB/NIH grants R01EB019036. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. We also thank Dr. Hanbin Mao at Kent State University and Dr. Soma Dhakal at Virginia Commonwealth University for allowing us to use their instrument for part of the project. We thank Hongran Yin for providing the urine assay data, Dr. Daniel Jasinski for generating Fig. 6 and contributing significantly to the preparation of the manuscript. We thank Kaiming Zhang and Wah Chiu for the preparation of the CryoEM images; Yuri L. Lyubchenko and Lyudmila S. Shlyakhtenko for the preparation of AFM images; Daniel Binzel, Lora McBride and Nicolas Burns for assistance in the preparation of the manuscript.
Footnotes
The Supporting Information is available on the ACS Publications website.
Figures of structure of 6WJ nanoparticles using Swiss PDB. Tables showing the nucleic acid oligo sequence used in in vivo studies as well as in single molecule studies.
Competing Interest Statement
P.G. is the consultant of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Biomedical Co. Ltd, and cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical Corporation LTD.
References
- 1.Seeman NC; Sleiman HF, DNA Nanotechnology. Nat. Rev. Mater 2017, 3, 17068. [Google Scholar]
- 2.Shu D; Shu Y; Haque F; Abdelmawla S; Guo P, Thermodynamically Stable RNA Three-Way Junction for Constructing Multifunctional Nanoparticles for Delivery of Therapeutics. Nat. Nanotechnol 2011, 6, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S; Jiang Q; Liu S; Zhang Y; Tian Y; Song C; Wang J; Zou Y; Anderson GJ; Han J-Y; Chang Y; Liu Y; Zhang C; Chen L; Zhou G; Nie G; Yan H; Ding B; Zhao Y, A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger In Vivo. Nat. Biotechnol 2018, 36, 258. [DOI] [PubMed] [Google Scholar]
- 4.Kershner RJ; Bozano LD; Micheel CM; Hung AM; Fornof AR; Cha JN; Rettner CT; Bersani M; Frommer J; Rothemund PWK; Wallraff GM, Placement and Orientation of Individual DNA Shapes on Lithographically Patterned Surfaces. Nat. Nanotechnol 2009, 4, 557–561. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y; Liu J, Functional DNA Nanotechnology: Emerging Applications of Dnazymes and Aptamers. Curr. Opin. Biotechnol 2006, 17, 580–588. [DOI] [PubMed] [Google Scholar]
- 6.Nam KT; Kim D-W; Yoo PJ; Chiang C-Y; Meethong N; Hammond PT; Chiang Y-M; Belcher AM, Virus-Enabled Synthesis and Assembly of Nanowires for Lithium Ion Battery Electrodes. Science 2006, 312, 885–888. [DOI] [PubMed] [Google Scholar]
- 7.Ohno H; Saito H, Chapter Six - RNA and RNP as Building Blocks for Nanotechnology and Synthetic Biology Progress in Molecular Biology and Translational Science; Academic Press: Massachusetts USA: 2016; Vol. 139, 165–185. [DOI] [PubMed] [Google Scholar]
- 8.Han D; Park Y; Kim H; Lee JB, Self-Assembly of Free-Standing RNA Membranes. Nat. Commun 2014, 5, 4367. [DOI] [PubMed] [Google Scholar]
- 9.Ko SH; Su M; Zhang C; Ribbe AE; Jiang W; Mao C, Synergistic Self-Assembly of RNA and DNA Molecules. Nat. Chem 2010, 2, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabow WW; Jaeger L, RNA Self-Assembly and RNA Nanotechnology. Acc. Chem. Res 2014, 47, 1871–1880. [DOI] [PubMed] [Google Scholar]
- 11.Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L, In Vitro Assembly of Cubic RNA-Based Scaffolds Designed in Silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S-J, RNA Folding: Conformational Statistics, Folding Kinetics, and Ion Electrostatics. Annu. Rev. Biophys 2008, 37, 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodson SA, Metal Ions and RNA Folding: A Highly Charged Topic with a Dynamic Future. Curr. Opin. Chem. Biol 2005, 9, 104–109. [DOI] [PubMed] [Google Scholar]
- 14.Kim D-N; Kilchherr F; Dietz H; Bathe M, Quantitative Prediction of 3D Solution Shape and Flexibility of Nucleic Acid Nanostructures. Nucleic Acids Res 2011, 40, 2862–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber G, Mesoscopic Model Parametrization of Hydrogen Bonds and Stacking Interactions of RNA from Melting Temperatures. Nucleic Acids Res. 2012, 41, e30–e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter AE; Turner DH; Kim J; Lyttle MH; Müller P; Mathews DH; Zuker M, Coaxial Stacking of Helixes Enhances Binding of Oligoribonucleotides and Improves Predictions of RNA Folding. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patra A; Paolillo M; Charisse K; Manoharan M; Rozners E; Egli M, 2′-Fluoro RNA Shows Increased Watson–Crick H-Bonding Strength and Stacking Relative to RNA: Evidence from Nmr and Thermodynamic Data. Angew. Chem. Int. Ed 2012, 51, 11863–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu Y; Yin H; Rajabi M; Li H; Vieweger M; Guo S; Shu D; Guo P, RNA-Based Micelles: A Novel Platform for Paclitaxel Loading and Delivery. J. Controlled Release 2018, 276, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JB; Hong J; Bonner DK; Poon Z; Hammond PT, Self-Assembled RNA Interference Microsponges for Efficient SiRNA Delivery. Nat. Mater 2012, 11, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S; Finzi L; Bustamante C, Direct Mechanical Measurements of the Elasticity of Single DNA Molecules by Using Magnetic Beads. Science 1992, 258, 1122–1126. [DOI] [PubMed] [Google Scholar]
- 21.Chiu H-C; Koh KD; Evich M; Lesiak AL; Germann MW; Bongiorno A; Riedo E; Storici F, RNA Intrusions Change DNA Elastic Properties and Structure. Nanoscale 2014, 6, 10009–10017. [DOI] [PubMed] [Google Scholar]
- 22.Luo Z; Cheng B; Cui S, Effects of Water on the Single-Chain Elasticity of Poly(U) RNA. Langmuir 2015, 31, 6107–6113. [DOI] [PubMed] [Google Scholar]
- 23.Lee G; Bratkowski MA; Ding F; Ke A; Ha T, Elastic Coupling between RNA Degradation and Unwinding by an Exoribonuclease. Science 2012, 336, 1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger DM; Bergs J; Kazemi S; Gohlke H, Target Flexibility in RNA–Ligand Docking Modeled by Elastic Potential Grids. ACS Med. Chem. Lett 2011, 2, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahsan A; Rudnick J; Bruinsma R, Soft Elasticity of RNA Gels and Negative Poisson Ratio. Phys. Rev. E 2007, 76, 061910. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C; Fu H; Yang Y; Zhou E; Tan Z; You H; Zhang X, The Mechanical Properties of RNA-DNA Hybrid Duplex Stretched by Magnetic Tweezers. Biophys. J 2019, 116, 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson DR; McIntosh DB; Stevens MJ; Rubinstein M; Saleh OA, Single-Stranded Nucleic Acid Elasticity Arises from Internal Electrostatic Tension. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jovin TM; McIntosh LP; Arndt-Jovin DJ; Zarling DA; Robert-Nicoud M; van de Sande JH; Jorgenson KF; Eckstein F, Left-Handed DNA: From Synthetic Polymers to Chromosomes. J. Biomol. Struct. Dyn 1983, 1, 21–57. [DOI] [PubMed] [Google Scholar]
- 29.Xu J; Fogleman EA; Craig SL, Structure and Properties of DNA-Based Reversible Polymers. Macromolecules 2004, 37, 1863–1870. [Google Scholar]
- 30.Lipfert J; Skinner GM; Keegstra JM; Hensgens T; Jager T; Dulin D; Köber M; Yu Z; Donkers SP; Chou F-C; Das R; Dekker NH, Double-Stranded RNA under Force and Torque: Similarities to and Striking Differences from Double-Stranded DNA. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 15408–15413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrero-Galán E; Fuentes-Perez ME; Carrasco C; Valpuesta JM; Carrascosa JL; Moreno-Herrero F; Arias-Gonzalez JR, Mechanical Identities of RNA and DNA Double Helices Unveiled at the Single-Molecule Level. J. Am. Chem. Soc 2013, 135, 122–131. [DOI] [PubMed] [Google Scholar]
- 32.Peng L; Wu C; You M; Han D; Chen Y; Fu T; Ye M; Tan W, Engineering and Applications of DNA-Grafting Polymer Materials. Chem. Sci 2013, 4, 1928–1938.23682309 [Google Scholar]
- 33.Lötters JC; Olthuis W; Veltink PH; Bergveld P, The Mechanical Properties of the Rubber Elastic Polymer Polydimethylsiloxane for Sensor Applications. J. Micromech. Mircoeng 1997, 7, 145–147. [Google Scholar]
- 34.Wu PD; Van Der Giessen E, On Improved Network Models for Rubber Elasticity and Their Applications to Orientation Hardening in Glassy Polymers. J. Mech. Phys. Solids 1993, 41, 427–456. [Google Scholar]
- 35.Kader MA; Bhowmick AK, Novel Thermoplastic Elastomers from Fluorocarbon Elastomer, Acrylate Rubber and Acrylate Plastics. Rubber Chem. Technol 2001, 74, 662–676. [Google Scholar]
- 36.Li H; Lee T; Dziubla T; Pi F; Guo S; Xu J; Li C; Haque F; Liang X-J; Guo P, RNA as a Stable Polymer to Build Controllable and Defined Nanostructures for Material and Biomedical Applications. Nano Today 2015, 10, 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gothelf KV, Lego-Like DNA Structures. Science 2012, 338, 1159–1160. [DOI] [PubMed] [Google Scholar]
- 38.Wang MD; Yin H; Landick R; Gelles J; Block SM, Stretching DNA with Optical Tweezers. Biophys. J 1997, 72, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangeol P; Côte D; Bizebard T; Legrand O; Bockelmann U, Probing DNA and RNA Single Molecules with a Double Optical Tweezer. Eur. Phys. J. E: Soft Matter Biol. Phys 2006, 19, 311–317. [DOI] [PubMed] [Google Scholar]
- 40.Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P, Enhancing Immunomodulation on Innate Immunity by Shape Transition among RNA Triangle, Square and Pentagon Nanovehicles. Nucleic Acids Res 2014, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chworos A; Severcan I; Koyfman AY; Weinkam P; Oroudjev E; Hansma HG; Jaeger L, Building Programmable Jigsaw Puzzles with RNA. Science 2004, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- 42.Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L, Design and Self-Assembly of SiRNA-Functionalized RNA Nanoparticles for Use in Automated Nanomedicine. Nat. Protoc 2011, 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiemann K; Rossi JJ, RNAi-Based Therapeutics–Current Status, Challenges and Prospects. EMBO Mol. Med 2009, 1, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu Y; Shu D; Haque F; Guo P, Fabrication of PRNA Nanoparticles to Deliver Therapeutic RNAs and Bioactive Compounds into Tumor Cells. Nat. Protoc 2013, 8, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaeger L; Westhof E; Leontis NB, Tectorna: Modular Assembly Units for the Construction of RNA Nano-Objects. Nucleic Acids Res. 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H; Zhang K; Pi F; Guo S; Shlyakhtenko L; Chiu W; Shu D; Guo P, Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting. Adv. Mater 2016, 28, 7501–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khisamutdinov EF; Jasinski DL; Li H; Zhang K; Chiu W; Guo P, Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater 2016, 28, 10079–10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C; Li H; Zhang K; Binzel DW; Yin H; Chiu W; Guo P, Photo-Controlled Release of Paclitaxel and Model Drugs from RNA Pyramids. Nano Res. 2019, 12, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A; Haque F; Pi F; Shlyakhtenko LS; Evers BM; Guo P, Controllable Self-Assembly of RNA Dendrimers. Nanomed. Nanotechnol. Biol. Med 2016, 12, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H; Wang H; Li Z; Shu D; Guo P, RNA Micelles for the Systemic Delivery of Anti-MiRNA for Cancer Targeting and Inhibition without Ligand. ACS Nano 2019, 13, 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hui Y; Yi X; Hou F; Wibowo D; Zhang F; Zhao D; Gao H; Zhao C-X, Role of Nanoparticle Mechanical Properties in Cancer Drug Delivery. ACS Nano 2019, 13, 7410–7424. [DOI] [PubMed] [Google Scholar]
- 52.Canton I; Battaglia G, Endocytosis at the Nanoscale. Chem. Soc. Rev 2012, 41, 2718–2739. [DOI] [PubMed] [Google Scholar]
- 53.Arvizo RR; Miranda OR; Moyano DF; Walden CA; Giri K; Bhattacharya R; Robertson JD; Rotello VM; Reid JM; Mukherjee P, Modulating Pharmacokinetics, Tumor Uptake and Biodistribution by Engineered Nanoparticles. PLoS ONE 2011, 6, e24374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anselmo AC; Zhang M; Kumar S; Vogus DR; Menegatti S; Helgeson ME; Mitragotri S, Elasticity of Nanoparticles Influences Their Blood Circulation, Phagocytosis, Endocytosis, and Targeting. ACS Nano 2015, 9, 3169–3177. [DOI] [PubMed] [Google Scholar]
- 55.Guo P; Liu D; Subramanyam K; Wang B; Yang J; Huang J; Auguste DT; Moses MA, Nanoparticle Elasticity Directs Tumor Uptake. Nat. Commun 2018, 9, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo S; Li H; Ma M; Fu J; Dong Y; Guo P, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther.-- Nucleic Acids 2017, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piao X; Yin H; Guo S; Wang H; Guo P, RNA Nanotechnology to Solubilize Hydrophobic Antitumor Drug for Targeted Delivery. Adv. Sci. (Weinheim, Ger.) 2019, 6, 1900951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin H; Xiong G; Guo S; Xu C; Xu R; Guo P; Shu D, Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther 2019, 27, 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longmire M; Choyke PL; Kobayashi H, Clearance Properties of Nano-Sized Particles and Molecules as Imaging Agents: Considerations and Caveats. Nanomedicine 2008, 3, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P, Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA 2013, 19, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H; Endrizzi JA; Shu Y; Haque F; Sauter C; Shlyakhtenko LS; Lyubchenko Y; Guo P; Chi Y-I, Crystal Structure of 3WJ Core Revealing Divalent Ion-Promoted Thermostability and Assembly of the Phi29 Hexameric Motor PRNA. RNA 2013, 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jasinski DL; Khisamutdinov EF; Lyubchenko YL; Guo P, Physicochemically Tunable Polyfunctionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties. ACS Nano 2014, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bustamante C; Marko J; Siggia E; Smith S, Entropic Elasticity of Lambda-Phage DNA. Science 1994, 265, 1599–1600. [DOI] [PubMed] [Google Scholar]
- 64.Santini GPH; Pakleza C; Cognet JAH, DNA Tri - and Tetra - Loops and RNA Tetra - Loops Hairpins Fold as Elastic Biopolymer Chains in Agreement with Pdb Coordinates. Nucleic Acids Res. 2003, 31, 1086–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi HJ; Boyce MC, Stress–Strain Behavior of Thermoplastic Polyurethanes. Mech. Mater 2005, 37, 817–839. [Google Scholar]
- 66.Treloar LRG, Stress-Strain Data for Vulcanized Rubber under Various Types of Deformation. Rubber Chem. Technol 1944, 17, 813–825. [Google Scholar]
- 67.Hanson DE, The Molecular Kink Paradigm for Rubber Elasticity: Numerical Simulations of Explicit Polyisoprene Networks at Low to Moderate Tensile Strains. J. Chem. Phys 2011, 135, 054902. [DOI] [PubMed] [Google Scholar]
- 68.Liu C; Qin H; Mather PT, Review of Progress in Shape-Memory Polymers. J. Mater. Chem 2007, 17, 1543–1558. [Google Scholar]
- 69.Lendlein A; Kelch S, Shape-Memory Polymers. Angew. Chem. Int. Ed 2002, 41, 2034–2057. [PubMed] [Google Scholar]
- 70.López-Manchado MA; Valentín JL; Carretero J; Barroso F; Arroyo M, Rubber Network in Elastomer Nanocomposites. Eur. Polym. J 2007, 43, 4143–4150. [Google Scholar]
- 71.Jonchhe S; Ghimire C; Cui Y; Sasaki S; McCool M; Park S; Iida K; Nagasawa K; Sugiyama H; Mao H, Binding of a Telomestatin Derivative Changes the Mechanical Anisotropy of a Human Telomeric G-Quadruplex. Angew. Chem. Int. Ed 2019, 58, 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marin-Gonzalez A; Vilhena JG; Perez R; Moreno-Herrero F, Understanding the Mechanical Response of Double-Stranded DNA and RNA Under Constant Stretching Forces Using All-Atom Molecular Dynamics. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 7049–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perera Rukshan T.; Fleming Aaron M.; Peterson Amberlyn M.; Heemstra Jennifer M.; Burrows Cynthia J.; White Henry S., Unzipping of a-Form DNA-RNA, A-Form DNA PNA, and B-Form DNA-DNA in the A-Hemolysin Nanopore. Biophys. J 2016, 110, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dudko OK; Hummer G; Szabo A Theory, Analysis, and Interpretation of Single-Molecule Force Spectroscopy Experiments. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 15755–15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stylianopoulos T, Epr-Effect: Utilizing Size-Dependent Nanoparticle Delivery to Solid Tumors. Ther. Delivery 2013, 4, 421–3. [DOI] [PubMed] [Google Scholar]
- 76.Jhaveri AM; Torchilin VP, Multifunctional Polymeric Micelles for Delivery of Drugs and SiRNA. Front. Pharmacol 2014, 5, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ellington AD; Szostak JW, In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 78.Roberts WG; Palade GE, Neovasculature Induced by Vascular Endothelial Growth Factor Is Fenestrated. Cancer Res 1997, 57, 765–72. [PubMed] [Google Scholar]
- 79.Guo P, The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shu D; Shu Y; Haque F; Abdelmawla S; Guo P, Thermodynamically Stable RNA Three-Way Junctions for Constructing Multifuntional Nanoparticles for Delivery of Therapeutics. Nat. Nanotechnol 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sindhwani S; Syed AM; Ngai J; Kingston BR; Maiorino L; Rothschild J; MacMillan P; Zhang Y; Rajesh NU; Hoang T; Wu JLY; Wilhelm S; Zilman A; Gadde S; Sulaiman A; Ouyang B; Lin Z; Wang L; Egeblad M; Chan WCW, The Entry of Nanoparticles into Solid Tumours. Nat. Mater 2020, 19, 566–575. [DOI] [PubMed] [Google Scholar]
- 82.Jasinski D; Haque F; Binzel DW; Guo P, Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo S; Xu C; Yin H; Hill J; Pi F; Guo P, Tuning the Size, Shape and Structure of RNA Nanoparticles for Favorable Cancer Targeting and Immunostimulation. Wiley Interdiscip Rev Nanomed. Nanobiotechnol 2020, 12, e1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin H; Wang H; Li Z; Shu D; Guo P, RNA Micelles for Systemic Delivery of Anti-MiRNA for Cancer Targeting and Inhibition without Ligand. ACS Nano 2018, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abdelmawla S; Guo S; Zhang L; Pulukuri S; Patankar P; Conley P; Trebley J; Guo P; Li QX, Pharmacological Characterization of Chemically Synthesized Monomeric PRNA Nanoparticles for Systemic Delivery. Mol. Ther 2011, 19, 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haque F; Shu D; Shu Y; Shlyakhtenko L; Rychahou P; Evers M; Guo P, Ultrastable Synergistic Tetravalent RNA Nanoparticles for Targeting to Cancers. Nano Today 2012, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P, Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA 2013, 19, 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haque F; Shu D; Shu Y; Shlyakhtenko LS; Rychahou PG; Mark Evers B; Guo P, Ultrastable Synergistic Tetravalent RNA Nanoparticles for Targeting to Cancers. Nano Today 2012, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jasinski DL; Li H; Guo P, The Effect of Size and Shape of RNA Nanoparticles on Biodistribution. Mol. Ther 2018, 26, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi HS; Liu W; Misra P; Tanaka E; Zimmer JP; Itty Ipe B; Bawendi MG; Frangioni JV, Renal Clearance of Quantum Dots. Nat. Biotechnol 2007, 25, 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J; Yu M; Zhou C; Zheng J, Renal Clearable Inorganic Nanoparticles: A New Frontier of Bionanotechnology. Mater. Today 2013, 16, 477–486. [Google Scholar]
- 92.Du B; Jiang X; Das A; Zhou Q; Yu M; Jin R; Zheng J, Glomerular Barrier Behaves as an Atomically Precise Bandpass Filter in a Sub-Nanometre Regime. Nat. Nanotechnol 2017, 12, 1096–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koirala D; Dhakal S; Ashbridge B; Sannohe Y; Rodriguez R; Sugiyama H; Balasubramanian S; Mao H, A Single-Molecule Platform for Investigation of Interactions between G-Quadruplexes and Small-Molecule Ligands. Nat. Chem 2011, 3, 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mao H; Luchette P, An Integrated Laser-Tweezers Instrument for Microanalysis of Individual Protein Aggregates. Sens. Actuators, B 2008, 129, 764–771. [Google Scholar]
- 95.Ghimire C; Park S; Iida K; Yangyuoru P; Otomo H; Yu Z; Nagasawa K; Sugiyama H; Mao H, Direct Quantification of Loop Interaction and Π–Π Stacking for G-Quadruplex Stability at the Submolecular Level. J. Am. Chem. Soc 2014, 136, 15537–15544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


