Abstract
Ampelocalamus naibunensis is one drooping bamboo with an important ornamental value endemic to Taiwan Island. To date, the genetic and genomic information of this species is little known. Here we characterized the complete chloroplast genome of A. naibunensis using genome skimming approach. The complete chloroplast genome is 139,860 bp, with a large single copy region (LSC) of 83,380 bp and a small single copy region (SSC) of 13,014 bp separated by a pair of inverted repeats (IRs) of 21,822 bp. The genome encodes a total of 129 genes, of which 111 are unique, containing 76 protein-coding genes, 4 ribosomal RNAs and 31 transfer RNAs. Sixteen distinct genes contain one or two introns, and the GC content of the cp genome is 38.9%. Phylogenomic analysis strongly supports the placement of A. naibunensis in the Chimonocalamus lineage (III), distantly related to A. calcareus (XI) within temperate woody bamboos.
Keywords: Chloroplast genome, Ampelocalamus naibunensis, genome skimming, phylogenomics
Ampelocalamus naibunensis (Hayata) T.H. Wen is stenochoric and endemic to southern Taiwan Island, with important ornamental value for its apically drooping appearances (Li et al. 2006). This species plays a crucial role in understanding the intriguing biogeographic pattern of Ampelocalamus, an obviously discontinuous distribution of mainland China-Taiwan Island-Hainan Island (Wu et al. 2010; Zhang et al. 2016a), as well as the spatial and temporal diversification of temperate woody bamboos (Arundinarieae) in Bambusoideae (Zhang et al. 2016b). In this study, we assembled and characterized the complete chloroplast (cp) genome of A. naibunensis via genome skimming approach (Straub et al. 2012), which will provide additional temp-spatially evolutionary information of the special forestry-adapted group of the grass family.
Total genomic DNA was extracted from fresh leaves of A. naibunensis grown in Kunming Botanic Garden in Kunming, Yunnan province of China. The voucher specimen was deposited at the Herbarium of Kunming Institute of Botany (accession number Zhang12318). Illumina paired-end (PE) library was constructed and sequenced (2 × 100 bp) from fragmented genomic DNA in Beijing Genomics Institute (BGI) in Shenzhen, China. The cp genome de novo assembly was performed using CLC Genomics Workbench v7.5 software (CLC Bio, Aarhus, Denmark). Obtained cp contigs (length >300 bp and sequence coverage >50) were retrieved and ordered by a BLAST search against the reference sequence of Phyllostachys edulis (HQ337796). The finished cp genome was annotated using DOGMA (Wyman et al. 2004), coupled with manual adjustments.
The assembled cp genome of A. naibunensis (GenBank accession KX372537) is 139,860 bp in size with high coverage (mean value 369.5), showing a typical quadripartite structure: two inverted repeats (IRs) of 21,822 bp separating one large single copy region (LSC) of 83,380 bp and one small single copy region (SSC) of 13,014 bp. A total of 129 genes are contained in the cp genome, of which 111 are unique, including 76 protein-coding genes, 4 ribosomal RNAs and 31 transfer RNAs. The overall GC content of the cp genome is 38.9%. One protein-coding gene (ycf3) contains two introns while another 9 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rps12, rps16) contain one intron each. Protein-coding regions contribute 42.4% of A. naibunensis cp genome which bears a high level of conservation in terms of gene content, gene order, GC content and the size of IRs compared with other already published cp genomes of Arundinarieae (Zhang et al. 2011).
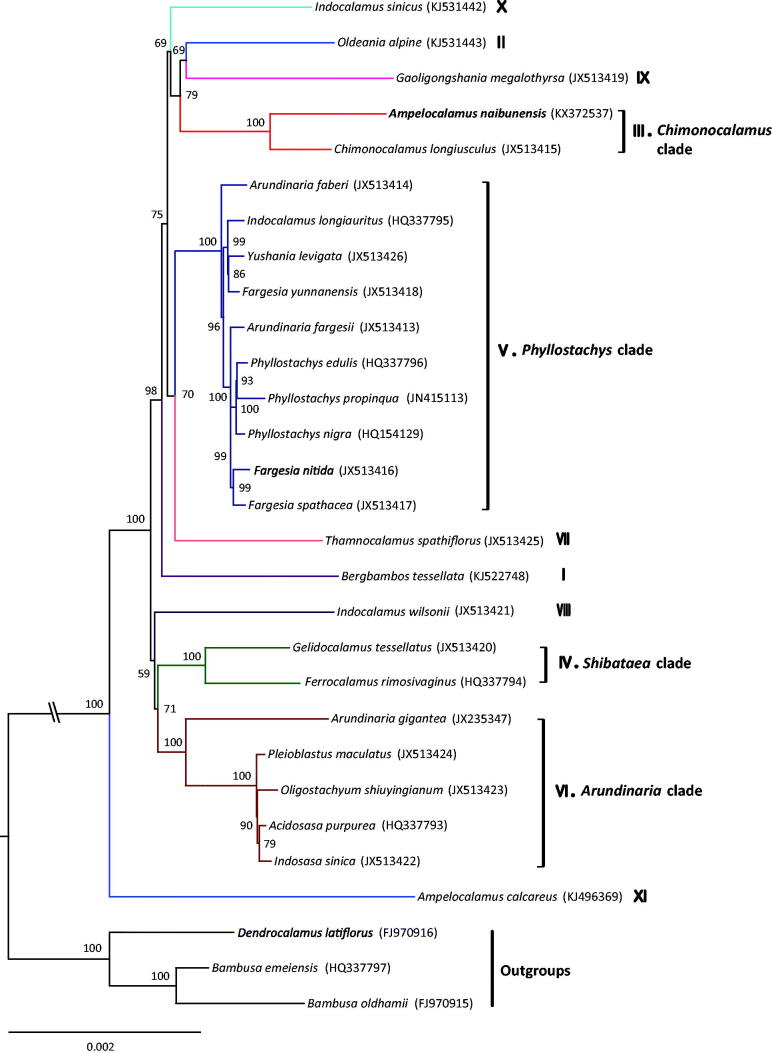
Phylogenomic analysis was performed based on the complete cp genomes of 29 bamboos using RAxML v.8.2.8 (Stamatakis 2014). We found that A. naibunensis clustered in the Chimonocalamus clade (III), being sister to C. longiusculus with high-support value (Figure 1). Ampelocalamus naibunensis is distantly related to A. calcareus, the earliest divergent lineage (XI) of Arundinarieae (Zhang et al. 2016b). The phylogenetic relationships of the 11 major lineages (I to XI) of Arundinarieae recovered here is congruent with Ma et al. (2014). The short internodes connected by long branches in the ML tree suggested a probable recent rapid radiation of Arundinarieae (Zhang et al. 2016b).
Figure 1.
Maximum likelihood tree inferred from 29 woody bamboo chloroplast genomes. Colored branches indicate the 11 Arundinarieae lineages (I to XI). The position of Ampelocalamus naibunensis is shown in bold. Values associated with nodes are bootstrapping supports.
Acknowledgement
The authors would like to thank Yuxiao Zhang for the help in sample collection.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work is supported by the Basic Science Fund (grant no. 2452016052) and Doctor Startup Fund (grant no. Z109021509) from Northwest A&F University
References
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA.. 2006. Bambuseae (Poaceae) In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. vol.22 Beijing and St. Louis: Science Press and Missouri Botanical Garden Press. [Google Scholar]
- Ma P-F, Zhang Y-X, Zeng C-X, Guo Z-H, Li D-Z.. 2014. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol. 63:933–950. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SC, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A.. 2012. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot. 99:349–364. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Sun H, Zhou ZK, Li DZ, Peng H.. 2010. Floristics of seed plants from China. Beijing: Science Press. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhang Y-J, Ma P-F, Li D-Z.. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One. 6:e20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-X, Ye X-Y, Yang H-M, Zhang X-Z, Wang P-Y, Li D-Z.. 2016a. New distribution records of two bamboo species in Yunnan, China with description of the inflorescence for Melocalamus yunnanensis (Poaceae, Bambusoideae). PhytoKeys. 62:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-Z, Zeng C-X, Ma P-F, Haevermans T, Zhang Y-X, Zhang L-N, Guo Z-H, Li D-Z.. 2016b. Multi-locus plastid phylogenetic biogeography supports the Asian hypothesis of the temperate woody bamboos (Poaceae: Bambusoideae). Mol Phylogenet Evol. 96:118–129. [DOI] [PubMed] [Google Scholar]