Abstract
Darkfin hind, Cephalopholis urodeta, belongs to the subfamily Epinephelinae. It is one of the most important fish species in coral-reef ecosystem. In this study, the complete mitochondrial (mt) genome of C. urodeta has been determined. It was 16,592 bp in length and contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes and 2 non-coding regions. The mitogenome sequence of C. urodeta shared 94% and 92% similarity to that of C. sonnerati and C.sexmaculata, respectively. Phylogenetic tree was made based on the concatenated sequences of 12 protein-coding genes on mtH-strand. All the results provide insights into the evolution in the subfamily Epinephelinae.
Keywords: Grouper, mitogenome, genetics
Groupers are bottom-associated fishes found in the tropical and subtropical waters of all oceans. Darkfin hind, Cephalopholis urodeta, is one of the common coral reef species of grouper found in outer reef areas, lagoons and back-reef areas and on the reef-top. It is a widespread species occurring at the tropical islands and shallow banks of the Indian and west-central Pacific Oceans, and the northern coast of Australia. Because of its small size (9–21 cm standard length), C. urodeta is not of much interest as a food fish (Heemstra & Randall 1993). However, C. urodeta shows subsistence commercial status (Tyler et al. 2009) and important ecological functions because it is one of the major predators feeding on a variety of fishes, crustaceans and cephalopods in coral-reef ecosystem (Randall & Brock 1960; Heemstra & Randall 1993; Pinault et al. 2014). Some confusions and disagreements remain puzzled on the classification and nomenclature of this species by morphological analysis (Allen & Steene 1988; Heemstra & Randall 1993).
In this study, three individuals of C. urodeta were obtained from Triton island (15°47′N 111°12′E) of China and species identifications were performed according to FAO Groupers of the World (Heemstra & Randall 1993). Dorsal muscle (Disposition number: ZJ201507A-C) were collected from frozen fishes. This study involving animals was carried out in accordance with the recommendations of “Animal Care and Ethical Committee, South China Sea Institute of Oceanology, Chinese Academy of Sciences.” Total genomic DNA was isolated from tissue samples of dorsal muscle using standard phenol-chloroform extraction and ethanol precipitation methods. The complete mitochondrial (mt) genome of C. urodeta was obtained with long PCR approach. Primers used were designed on the basis of aligned mitogenome sequences of C. sonnerati (KC593378.1), C. argus (KC593377.1), C. boenak (KC537759.1) and C. sexmaculata (KJ469385.1).
The complete mtDNA sequence of C. urodeta (GenBank accession number: KU891818) was 16,592 bp in length, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two non-coding regions: origin of light-strand replication (OL) and control region (CR or D-loop) (Table 1). Most of the genes were encoded on the heavy strand (H strand) except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu and tRNAPro), which are encoded on the L-strand. All genes showed the typical gene arrangement conforming to the vertebrate consensus (Johansen et al. 1990; Boore 1999). Sequence overlaps were showed between protein-coding genes, including ATP8-ATP6, ATP6-COIII, NDL4-ND4, and ND5-ND6, and/or tRNA genes, such as tRNAIle- tRNAGln, ND2- tRNATrp, COIII- tRNAGly, and tRNATyr- tRNAPro. The 40 bp fragment of OL, as in most vertebrates, overlapped the tRNACys gene by 1 bp and was located in a cluster of five tRNA genes (WANCY region; Table 1) between the tRNAAsn and tRNACys. The other non-coding region CR was bound by tRNAPro and tRNAPhe. Overall base composition of the mitogenome was estimated to be 29.47% A, 28.36% C, 15.99% G and 26.18% T, respectively, with a high A + T content (55.64%), indicating an obvious anti-guanine bias commonly observed in fishes (Cantatore et al. 1994; Wang et al. 2008). The mitogenome sequence of C. urodeta showed 94% and 92% identity to that of C. sonnerati and C. sexmaculata, respectively.
Table 1.
Characteristics of the mtgenome of C. urodeta.
| Size |
Codon |
||||||
|---|---|---|---|---|---|---|---|
| Locus | Nucleotide (Position) | Amino acid | Start | Stop | Anti-codon | Intergenic nucleotidea | Strandb |
| tRNAPhe | 70 (1–70) | GAA | 0 | H | |||
| 12S rRNA | 957 (71–1027) | 0 | H | ||||
| tRNAVal | 71 (1028–1098) | TAC | 1 | H | |||
| 16S rRNA | 1715 (1100–2814) | 1 | H | ||||
| tRNALeu(UUR) | 75 (2815–2889) | TAA | 0 | H | |||
| ND1 | 975 (2890–3864) | 324 | ATG | TAA | 6 | H | |
| tRNAIle | 70 (3871–3940) | GAT | −2 | H | |||
| tRNAGln | 71 (4009–3939) | TTG | 0 | L | |||
| tRNAMet | 69 (4010–4078) | CAT | 0 | H | |||
| ND2 | 1047 (4079–5124) | 348 | ATG | TA- | 0 | H | |
| tRNATrp | 71 (5125–5195) | TCA | 1 | H | |||
| tRNAAla | 69 (5265–5197) | TGC | 0 | L | |||
| tRNAAsn | 73 (5338–5266) | GTT | 0 | L | |||
| OL | 40 (5339–5378) | −1 | – | ||||
| tRNACys | 68 (5445–5378) | GCA | 0 | L | |||
| tRNATyr | 71 (5516–5446) | GTA | 1 | L | |||
| COI | 1551 (5518–7068) | 516 | GTG | TAG | 0 | H | |
| tRNASer(UCN) | 71 (7139–7069) | TGA | 3 | L | |||
| tRNAAsp | 73 (7143–7215) | GTC | 8 | H | |||
| COII | 691 (7224–7914) | 230 | ATG | T– | 0 | H | |
| tRNALys | 73 (7915–7987) | TTT | 1 | H | |||
| ATP8 | 168 (7989–8156) | 55 | ATG | TAA | −10 | H | |
| ATP6 | 684 (8147–8830) | 227 | CTG | TAA | −1 | H | |
| COIII | 786 (8830–9614) | 261 | ATG | TA- | 0 | H | |
| tRNAGly | 72 (9615–9686) | TCC | 0 | H | |||
| ND3 | 349 (9687–10,035) | 116 | ATG | T– | 0 | H | |
| tRNAArg | 69 (10,036–10,104) | TCG | 0 | H | |||
| ND4L | 297 (10,105–10,401) | 98 | ATG | TAA | −7 | H | |
| ND4 | 1381 (10,395–11,775) | 460 | ATG | T– | 0 | H | |
| tRNAHis | 70 (11,776–11,845) | GTG | 0 | H | |||
| tRNASer(AGY) | 75 (11,846–11,920) | GCT | 9 | H | |||
| tRNALeu(CUN) | 73 (11,930–12,002) | TAG | 0 | H | |||
| ND5 | 1839 (12,003–13,841) | ATG | TAA | −4 | H | ||
| ND6 | 522 (14,359–13,838) | 173 | ATG | TAA | 0 | L | |
| tRNAGlu | 69 (14,428–14,360) | TTC | 4 | L | |||
| Cyt b | 1141 (14,433–15,573) | 377 | ATG | T– | 0 | H | |
| tRNAThr | 73 (15,574–15,646) | TGT | −1 | H | |||
| tRNAPro | 70 (15,715–15,646) | TGG | 0 | L | |||
| D-loop | 877 (15,716–16,592) | – | |||||
aNumbers correspond to the nucleotides separating different genes. Negative numbers indicate overlapping nucleotides between adjacent genes.
bH and L indicate genes transcribed on the heavy and light strands, respectively.
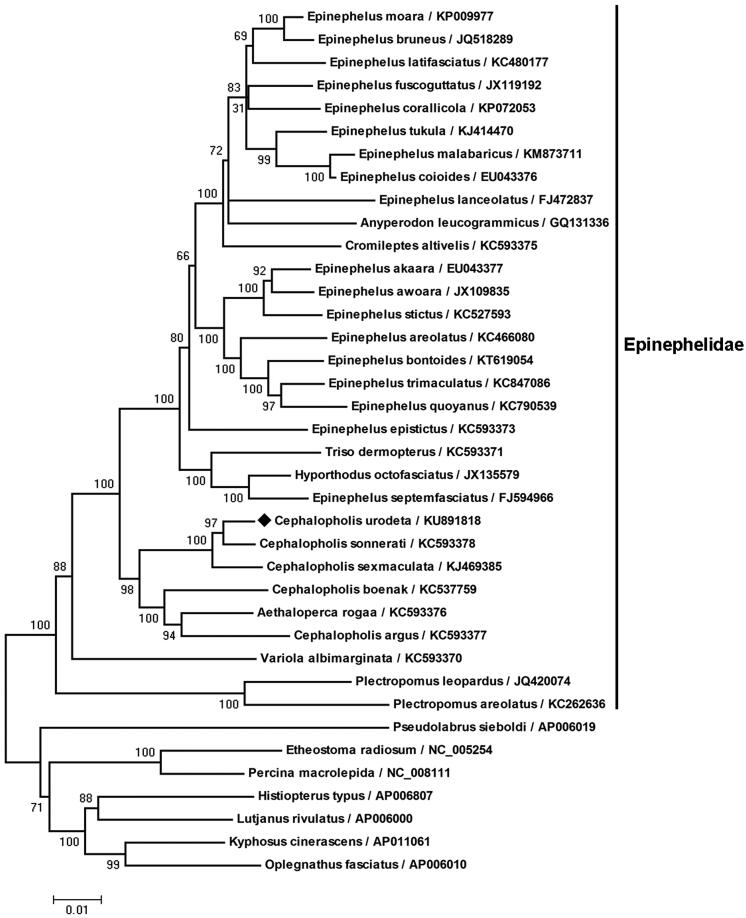
Most of the grouper mtprotein-coding genes began with the typical start codon ATG. As in many other metazoans (Wolstenholme 1992), the COI gene began with GTG in C. urodeta. Different from most other teleosts and basal groupers (Craig & Hastings 2007; Zhuang et al. 2013), CTG was the start codon of the ATP6 gene in C. urodeta (Table 1). The protein-coding genes COII, ND3, ND4 and Cyt b were all terminated with the incomplete stop codon T–, while ND2 and COIII were TA- (Table 1). It was completed with the addition of 3′ adenine residues to the mRNA by post-transcriptional polyadenylation (Ojala et al. 1981; Coucheron et al. 2011). The pattern of codon usage in the C. urodeta mtDNA is shown in Table 2. There were 3807 codons for all the protein-coding genes after excluding the incomplete stop codons. The concatenated sequences of 12 protein-coding genes on mtH-stand were aligned with codon constraint using Clustal X (http://www.ebi.ac.uk/clustalW/). Phylogenetic tree (Figure 1) was constructed according to the alignment of amino acid sequences with MEGA 4.0 (http://megasoftware.net).
Table 2.
Codon usage of the protein-coding genes in C. urodeta mtgenome.
| Amino acid | Codon | No. | % | Amino acid | Codon | No. | % |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 80 | 2.10 | Stop | UAA | 6 | 0.21 |
| UUC | 158 | 4.15 | UAG | 1 | 0.03 | ||
| Leu | UUA | 94 | 2.47 | His | CAU | 30 | 0.79 |
| UUG | 12 | 0.32 | CAC | 76 | 2.00 | ||
| CUU | 137 | 3.60 | Gln | CAA | 93 | 2.44 | |
| CUC | 126 | 3.31 | CAG | 7 | 0.18 | ||
| CUA | 241 | 6.33 | Asn | AAU | 42 | 1.10 | |
| CUG | 33 | 0.87 | AAC | 78 | 2.05 | ||
| Ile | AUU | 144 | 3.78 | Lys | AAA | 73 | 1.92 |
| AUC | 130 | 3.41 | AAG | 4 | 0.11 | ||
| Met | AUA | 100 | 2.63 | Asp | GAU | 31 | 0.81 |
| AUG | 63 | 1.65 | GAC | 49 | 1.29 | ||
| Val | GUU | 58 | 1.52 | Glu | GAA | 82 | 2.15 |
| GUC | 56 | 1.47 | GAG | 13 | 0.34 | ||
| GUA | 96 | 2.52 | Cys | UGU | 13 | 0.34 | |
| GUG | 13 | 0.34 | UGC | 14 | 0.37 | ||
| Ser | UCU | 39 | 1.02 | Trp | UGA | 104 | 2.74 |
| UCC | 74 | 1.94 | UGG | 13 | 0.34 | ||
| UCA | 68 | 1.79 | Arg | CGU | 12 | 0.32 | |
| UCG | 7 | 0.18 | CGC | 10 | 0.26 | ||
| Pro | CCU | 48 | 1.26 | CGA | 47 | 1.23 | |
| CCC | 88 | 2.31 | CGG | 9 | 0.24 | ||
| CCA | 75 | 1.97 | Ser | AGU | 4 | 0.11 | |
| CCG | 7 | 0.18 | AGC | 46 | 1.21 | ||
| Thr | ACU | 46 | 1.21 | AGA | *** | — | |
| ACC | 115 | 3.02 | AGG | *** | — | ||
| ACA | 129 | 3.39 | Gly | GGU | 53 | 1.39 | |
| ACG | 13 | 0.34 | GGC | 74 | 1.94 | ||
| Ala | GCU | 84 | 2.21 | GGA | 88 | 2.31 | |
| GCC | 128 | 3.36 | GGG | 27 | 0.71 | ||
| GCA | 130 | 3.41 | NNAa | 1426 | 37.49 | ||
| GCG | 7 | 0.18 | NNTa | 860 | 22.58 | ||
| Tyr | UAU | 39 | 1.02 | NNCa | 1292 | 33.92 | |
| UAC | 70 | 1.84 | NNGa | 229 | 6.01 |
A total of 3807 codons were analyzed excluding the incomplete stop codons.
Amount and percentages of codons with the 3rd site nucleotide composition of A, T, C, G.
*** the stop code AGA and/or AGG (instead of Ser) was not detected.
Figure 1.
Phylogenetic , tree of C. urodeta and other fishes in suborder Percoidei. Phylogenetic tree was constructed according to the alignment of amino acid sequences of 12 protein-coding genes on mtH-strand by the neighbour-jointing method within MEGA 4.0 performing 1000 replications of bootstrap. The bootstrap values were indicated at the nodes of the tree. NCBI RefSeq or GenBank accession number of each species was listed on the right of the species name. Cephalopholis urodeta was clustered into the branch of family Epinephelidae.
The 12S rRNA and 16S rRNA genes lied between tRNAphe and tRNAVal, and tRNAVal and tRNALeu (UUR), respectively. A moderate nucleotide compositional bias, A (32.71%) >C (25.11%) >T (21.22%)>G (20.96%), was found in rRNA genes of C. urodeta. The tRNA genes ranged in size from 68 to 75 bp. Two forms of tRNALeu (UUR and CUN) and tRNASer (UCN and AGY) were contained in the mtgenome of C. urodeta (Table 1). Most tRNAs could be folded into the typical clover-leaf secondary structure by tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/). However, tRNASer (AGY) was found to lack the entire dihydrouridine (DHU) arm, reducing its secondary structure to a ‘truncated cloverleaf’. Similar phenomena have been reported in groupers (Zhuang et al. 2013) and most metazoans (Garey & Wolstenholme 1989). Aligning with sequences from other grouper species, CR of C. urodeta contained with three domains: the extended termination associated sequences (ETAS), central conserved domain (CCD), and conserved sequence blocks (CSB). The motif-TACAT and reversed motif-ATGTA were observed in the ETAS domains. Both motifs could form stable hairpin loops which presumably act as sequence-specific signals for termination of mtDNA replication (Saccone et al. 1991). All the data would contribute to the genetic conservation, species identification and phylogeny analysis of Epinephelinae.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the Natural Science Foundation of China [grant number U1301232], Major Science and Technology Program of Hainan Province [grant number ZDZX2013014] and the National Key Technology Support Program [grant number 2014BAC01B03].
References
- Allen GR, Steene RC.. 1988. Fishes of Christmas Island, Indian Ocean. Christmas Island, Indian Ocean, Australia: Christmas Island Natural History Association; pp. 197. [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P, Roberti M, Pesole G, Ludovico A, Milella F, Gadaleta MN, Saccore G.. 1994. . Evolutionary analysis of cytochrome b sequences in some Perciformes: evidence for a slower rate of evolution than in mammals. J Mol Evol. 39:589–597. [DOI] [PubMed] [Google Scholar]
- Coucheron DH, Nymark M, Breines R, Karlsen BO, Andreassen M.. 2011. . Characterization of mitochondrial mRNAs in codfish reveals unique features compared to mammals. Curr Genet. 57:213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig MT, Hastings PA.. 2007. A molecular phylogeny of the groupers classification of the Epinephelini. Ichthyol Res. 54:1–17. [Google Scholar]
- Garey JR, Wolstenholme DR.. 1989. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine are replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 28:374–387. [DOI] [PubMed] [Google Scholar]
- Heemstra PC, Randall JE.. 1993. FAO species catalogue: vol. 16. Groupers of the world (Family Serranidae, Subfamily Epinephelinae) In: Carpenter KE, Sommer C, eds. FAO fisheries synopsis. Rome: FAO; p. 61–62. [Google Scholar]
- Johansen S, Guddal PH, Johansen T.. 1990. Organization of the mitochondrial genome of Atlantic cod, Gadus morhua. Nucleic Acids Res. 18:411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G.. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474. [DOI] [PubMed] [Google Scholar]
- Pinault M, Bissery C, Gassiole G, Magalon H, Quod JP, Galzin R.. 2014. Fish community structure in relation to environmental variation in coastal volcanic habitats. J Exp Marine Biol Ecol. 460:62–71. [Google Scholar]
- Randall JE, Brock VE.. 1960. Observations on the ecology of epinepheline and lutjanid fishes of the Society Islands, with emphasis on food habits. Trans Am Fish Soc. 891:9–16. [Google Scholar]
- Saccone C, Pesole G, Sbisá E.. 1991. The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J Mol Evol. 33:83–91. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S.. 2007. . MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- Tyler EHM, Speight MR, Henderson P, Manica A.. 2009. Evidence for a depth refuge effect in artisanal coral reef fisheries. Biol Conserv. 142:652–667. [Google Scholar]
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S.. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Qu M, Zhang X, Ding S.. 2013. A comprehensive description and evolutionary analysis of 22 grouper (Perciformes, Epinephelidae) mitochondrial genomes with emphasis on two novel genome organizations. PLoS One. 8:e73561. [DOI] [PMC free article] [PubMed] [Google Scholar]