Abstract
The complete chloroplast genome sequence of Saussurea polylepis, one of vulnerable and endemic species of Korea, was determined. The genome size was 152,488 bp in length with 37.7% GC content. It included a pair of inverted repeats (IRa and IRb) of 25,191 bp, which were separated by small single copy (SSC: 18,689 bp) and large single copy (LSC: 83,417 bp) regions. The cp genome contained 113 genes, including 80 protein-coding genes, 29 tRNA genes, and four rRNA genes. Phylogenetic analysis of the combined 80 protein coding genes and four rRNA genes showed that S. polylepis was closely related to S. chabyoungsanica.
Keywords: Chloroplast genome, Saussurea polylepis, Korean endemic, vulnerable species
Saussurea (ca. 400 species) is one of the largest genera in the tribe Cardueae and is widely distributed throughout the Holarctic (Lipschitz 1979). The most comprehensive infrageneric classification system was proposed by Lipschitz (1979), recognizing six subgenera and 20 sections. In the most recent floristic treatment of Korea, approximately 34 species have been recognized (Im 2007). Of ca. 34 species, nearly 40% (13 species) are endemic to Korea, mostly occurring as small-sized populations in isolated regions of high elevation mountains and small islands. While most species occur in the backbone of Korean mountain ranges known as Baekdudaegan, S. polylepis is restricted to small clustered islands in southwestern part of the Korean peninsula. Since only a few individuals are found in a total of 10 locations, it is recognized as vulnerable species [VU B2ab (iii, iv)] in the Korean Red List (National Institute of Biological Resources 2014).
Here, we report the complete chloroplast genome of S. polylepis. The plant materials were sampled from Hongdo Island, Korea. The voucher specimen has been deposited at the Sungkyunkwan University Herbarium (SKK044232). Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA), following the instructions from the manufacturer. Genomic sequencing by an Illumina Miseq (Illumina Inc., San Diego, CA) platform was performed and chloroplast genome was assembled using SPAdes 2.425 and CLC Genomics Workbench v.5.5.1 (CLC Bio, Aarhus, Denmark). The chloroplast genome of S. polylepis was annotated using the DOGMA (Wyman et al. 2004) and CpGAVAS (Liu et al. 2012). The tRNA regions were confirmed using tRNAscan-SE with default setting (Schattner et al. 2005). Annotation of the start/stop codons of protein-coding genes was done using BLASTX, Geneious v.10.2.2. (Biomatters Ltd., Auckland, New Zealand), and then manually corrected for intron/exon boundaries. Maximum-likelihood analysis, including nine species from tribe Cynareae and 11 species representing seven tribes of the family Asteraceae, was conducted based on concatenated 84 chloroplast coding genes using IQ-TREE v.1.4.2 (Nguyen et al. 2015) with 1000 bootstrap (BS) replications.
The complete chloroplast genome of S. polylepis (MF695711) was 152,488 bp with 37.7% GC content and contained two inverted repeat regions (IRa and IRb) of 25,191 bp, separating a large single copy (LSC) region of 83,417 bp and a small single copy (SSC) region of 18,689 bp. The chloroplast genome contained 113 genes, including 80 protein-coding genes, 29 tRNA genes, and four rRNA genes. Nineteen genes, including seven tRNA genes and four rRNA genes, were duplicated in the IR regions. The structure, gene order, gene content, and GC content of the S. polylepis are similar to those of Cynareae chloroplast genomes (Curci et al. 2016; Lu et al. 2016; Cheon et al. 2017).
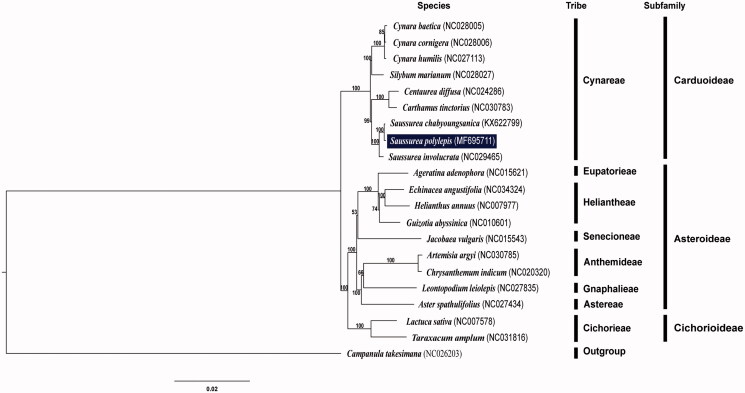
The phylogenetic analysis of major tribes of Asteraceae showed that tribe Cynareae, including three species of Saussurea, were monophyletic (100% BS) (Figure 1) and S. polylepis was most closely related to congeneric species S. chabyoungsainca in Korea. A further detailed work is necessary to elucidate phylogenetic relationships among major lineages of highly complex and specious genus Saussurea.
Figure 1.
Maximum-likelihood tree based on 80 protein-coding and four rRNA genes from 20 representative species of Asteraceae. Campanula takesimana (Campanulaceae) was used as an outgroup and the bootstrap support values >50% are shown at the branches.
Acknowledgements
The authors thank the Korean National Park Service for issuing a special permit to collect S. polylepis. The authors also thank Joon-Mo Lee for technical help during the assembly of chloroplast genome.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
References
- Cheon KS, Kim HJ, Han JS, Kim KA, Yoo KO.. 2017. The complete chloroplast genome sequence of Saussurea chabyoungsanica (Asteraceae), an endemic to Korea. Conserv Genet Resour. 9:51–53. [Google Scholar]
- Curci PL, De Paola D, Sonnante G.. 2016. Development of chloroplast genomic resources for Cynara. Mol Ecol Resour. 16:562–573. [DOI] [PubMed] [Google Scholar]
- Im HT. 2007. Saussurea DC In: Park CW, editor. The genera of vascular plants of Korea. Seoul (South Korea): Academy Publishing Co; p. 982–989. [Google Scholar]
- Lipschitz SJ. 1979. Genus Saussurea DC. (Asteraceae). Lenipopoli: Lenipopoli Science Press. [Google Scholar]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X.. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Shen Q, Yang J, Wang B, Song C.. 2016. The complete chloroplast genome sequence of Safflower (Carthamus tinctorius L.). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3351–3353. [DOI] [PubMed] [Google Scholar]
- National Institute of Biological Resources 2014. Korean Red List of Threatened Species. 2nd ed Incheon (South Korea): Jisungsa Publisher. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]