Abstract
Hosta capitata (Koidz.) Nakai is an herbaceous perennial plant belonging to the Asparagaceae family and has become a popular ornamental plant. In this study, the chloroplast genome sequence of H. capitata was completed by de novo assembly with whole genome sequence data. The chloroplast genome of H. capitata is 156,416 bp in length, which is composed of a large single-copy (LSC) of 84,788 bp, a small single-copy (SSC) of 18,206 bp, and a pair of inverted repeat regions (IRa and IRb) of 26,711 bp, as four distinct parts. In total, 114 genes were identified including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis revealed that H. capitata has a close relationship with other Hosta species, H. minor and H. ventricosa, but is farther than the distance between them.
Keywords: Hosta capitata (Koidz.) Nakai, chloroplast genome, phylogenetic analysis
Hosta capitata (Koidz.) Nakai is an herbaceous perennial plant belonging to the Asparagaceae family, which is seen in southern Korea and southwestern Japan (Fuhita 1976; Chung et al. 1991). Like other Hosta plants, H. capitata has become a popular ornamental plant as a shade-tolerant garden plant with characteristics of ovate leaves, purple flowers, and smaller size than others (Chung et al. 1991; Ryu et al. 2002; Bożek et al. 2015). In spite of its horticultural and economic importance, there is few genetic and genomic study for breeding of this plant. In this study, we completed the complete chloroplast genome sequence assembly of H. capitata using whole genome shotgun sequences and analyzed its phylogenetic relationship with other Asparagaceae species to provide useful genetic information for breeding.
To complete the chloroplast genome sequence, the total genomic DNA was extracted from leaves of H. capitata provided by HanTaek Botanical Garden, Yongin, Korea. The extracted DNA was implemented for the whole genome sequencing with the Illumina MiSeq platform (Illumina, San Diego, CA). The high-quality paired-end reads of 1.5 Gbp were obtained and assembled using CLC genome assembler (ver. 4.6, CLC Inc, Aarhus, Denmark), following dnaLCW method (Kim et al. 2015a, 2015b). After de novo assembly, four sequence contigs representing chloroplast genome were selected, ordered and combined into a single draft sequence by comparing with H. minor (KX822777) chloroplast genome sequence as a reference (Kim et al. 2012).
The complete genome of H. capitata (GenBank Accession No. MH581151) was 156,416 bp long, which consisted of a large single-copy (LSC) of 84,788 bp, a small single-copy (SSC) of 18,206 bp, and a pair of inverted repeat regions (IRa and IRb) of 26,711 bp. In the genome, a total of 114 genes were identified, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes, through Ge-Seq annotation and BLAST tools (Tillich et al. 2017).
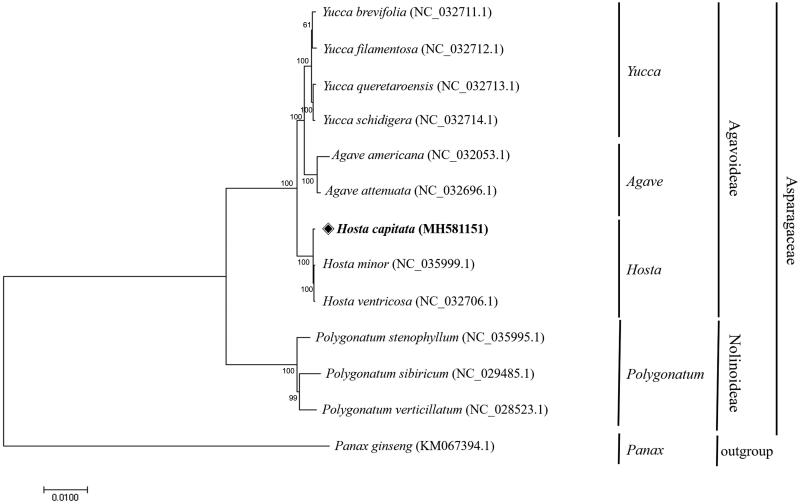
After multiple alignment of the chloroplast genome of H. capitata with those of 11 species belonging to the Asparagaceae family and Panax ginseng as an outgroup using MAFFT 7.0 (Katoh and Standley 2013), phylogenetic analysis was carried out based on the aligned sequences for a neighbor-joining tree by maximum composite likelihood model with 1000 of bootstrap replications of MEGA 7.0 (Kumar et al. 2016) (Figure 1). The phylogenetic tree showed that the Agavoideae and Nolinoideae subfamilies were divided and Agavoideae was further subgrouped into three genera, Yucca, Agave, and Hosta, respectively. To be specific, H. capitata was located together with H. minor and H. ventricosa, but is farther than the distance between the two Hosta plants.
Figure 1.
The phylogenetic tree was constructed using chloroplast genome sequences of 12 species belonging to the Asparagaceae family and Panax ginseng as an outgroup. On the MEGA 7.0 software, a neighbor-joining tree was constructed using maximum composite likelihood model with 1000 of bootstrap replications.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bożek M, Strzałkowska-Abramek M, Denisow B. 2015. Nectar and pollen production and insect visitation on ornamentals from the genus hosta tratt.(Asparagaceae). J Apic Sci. 59(2):115–125. [Google Scholar]
- Chung MG, Hamrick J, Jones SB Jr, Derda GS. 1991. Isozyme variation within and among populations of hosta (Liliaceae) in Korea. Syst Bot. 16(4):667–684. [Google Scholar]
- Fuhita N. 1976. Genus Hosta (Liliaceae) in Japan. Acta Phytotaxon Geobot Kyoto. 27:66–96. [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-K, Kim JS, Kim J-H. 2012. The phylogenetic relationships of asparagales in Korea based on five plastid DNA regions. J Plant Biol. 55(4):325–341. [Google Scholar]
- Kim K, Lee S-C, Lee J, Lee HO, Joh HJ, Kim N-H, Park H-S, Yang T-J. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45s nrdnas within panax ginseng species. PloS One. 10(6):e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of oryza aa genome species. Scient Reports. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K-H, Park M-H, Lee J-S. 2002. Occurrence of mosaic disease of hosta plane caused by hosta virus x. Plant Path J. 18(6):313–316. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. Geseq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]