Abstract
A Citrus hybrid, Hallabong mandarin, is a major Citrus tree largely cultivated in Korea. The complete chloroplast genome sequence of Hallabong mandarin was characterized by de novo assembly using whole genome next generation sequences. The chloroplast genome was 160 703 bp in length and separated into four distinct regions such as large single copy region (87 922 bp), small single copy region (18 801 bp) and a pair of inverted repeat regions (26 990 bp). The genome contained a total of 114 genes including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Phylogenetic inference using chloroplast genome sequences revealed that Hallabong mandarin was close to Citrus aurantiifolia (Omani lime) and Citrus sinensis (sweet orange).
Keywords: Chloroplast, citrus, genome sequence, Hallabong
Introduction
The Citrus genus in the Rutaceae family consists of important cultivated fruit trees worldwide, such as mandarins, oranges, lemons and lime (Zhang & Mabberley 2008). Because economic importance and consumers’ demands, various hybrids with desirable traits have been bred by crossing between Citrus species (Cameron & Frost 1968; Penjor et al. 2013). Among the hybrids, Hallabong mandarin, also called Shiranuhi in Japan, was bred by crossing between ‘Kiyomi’ (Citrus unshiu Marcov.×C. sinensis Osbeck) and ‘Nakano No.3’ Ponkan (C. reticulata Blanco) in Japan and produces almost seedless fruits (Matsumoto 2001). At present, Hallabong mandarin is one of the major Citrus hybrids cultivated and sold in Korea, owing to its unique taste and fragrance (Song et al. 2005; Kim et al. 2006). Although genetic resources for nuclear and chloroplast genomes are available for several Citrus and related species in the Rutaceae family (Bausher et al. 2006; Xu et al. 2013; Su et al. 2014, Wu et al. 2014; Lee et al. 2015), currently available genetic resources for Citrus hybrids are very limited. Therefore, we characterized the complete chloroplast genome sequence of a Citrus hybrid, Hallabong mandarin, to provide basic genetic resource for Citrus hybrids with other species in the Citrus genus.
Total genomic DNA was isolated from leaves sampled from a mandarin farm in Seogwipo, Jeju Special Self-Governing Province, Korea. An Illumina paired-end (PE) genomic library with 300-bp insert was constructed and sequenced using an Illumina HiSeq platform by Seeders Inc (http://www.seeders.co.kr/), Deajon, Korea. PE reads of 3 Gb were obtained and assembled by a CLC genome assembler (ver. 4.06 beta, CLC Inc, Aarhus, Denmark), as mentioned in Kim et al. (2015a, 2015b). The representative chloroplast contigs were collected, ordered and merged into a single draft sequence, through comparison with the chloroplast sequence of C. sinensis (DQ864733, Bausher et al. 2006) as a reference. The draft sequence was confirmed and manually corrected by PE read mapping. The genes in the chloroplast genome were annotated using the DOGMA program (Wyman et al. 2004) and BLAST searches.
The complete chloroplast genome of Hallabong mandarin (GenBank accession KU170678) was a circular molecule of 160 703 bp in length, which consisted of four distinct regions such as large single copy region of 87 922 bp, small single copy region of 18 801 bp and a pair of inverted repeat regions of 26 990 bp. Overall GC contents of chloroplast genomes was approximately 38.4%. The chloroplast genome contained a total of 114 genes including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The complete genome sequence showed 98.6% and 98.9% similarity with those of C. aurantiifolia (Omani lime) and C. sinensis (sweet orange), respectively.
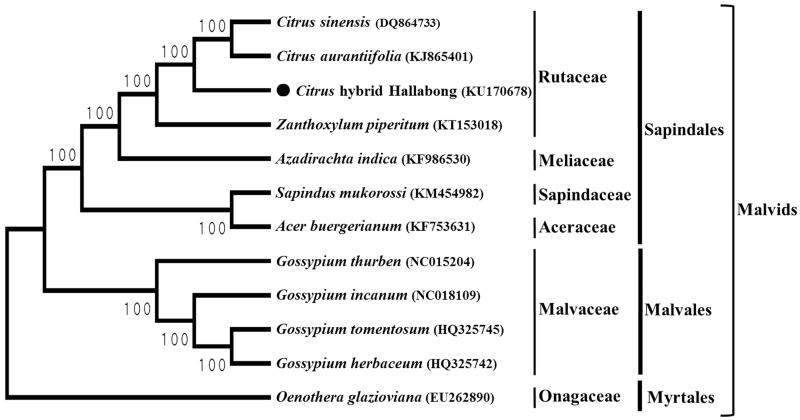
Phylogenetic analysis was carried out using the complete chloroplast genome sequence of Hallabong mandarin with those of 11 species in Malvids by a maximum likelihood (ML) analysis of MEGA 6.0 (Tamura et al. 2013). The phylogenetic tree showed that Hallabong mandarin was grouped to other species in the Rutaceae family and placed most closely to two Cirus species, C. aurantiifolia and C. sinensis, as expected (Figure 1).
Figure 1.
ML phylogenetic tree of Hallabong mandarin with 11 species in Malvids based on complete chloroplast genome sequences. Numbers in the nodes are bootstrap values from 1000 replicates. The chloroplast sequence of Oenothera glazioviana (large-flowered evening primrose) was set as an outgroup within Malvids.
Declaration of interest
This research was supported by Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-3-SB110, 213003-04-3-SBV10), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS).
References
- Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H.. 2006. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var “Ridge Pineapple”: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JW, Frost HB.. 1968. Genetics, breeding, and nucellar embryony In: Reuther W, Batchelor LD, Webber HJ, editors. The citrus industry. Vol.2 Berkeley: University of California Press; p. 325–370. [Google Scholar]
- Kim HS, Lee SH, Koh JS.. 2006. Physicochemical properties of Hallabong Tangor (Citrus Kiyomi × ponkan) cultivated with heating. Korean J Food Preserv. 13:611–615. [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ.. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10: e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee HJ, Kim K, Lee SC, Sung SH, Yang TJ.. 2015. The complete chloroplast genome sequence of Zanthoxylum Piperitum. Mitochondrial DNA Early Online. 1–2. DOI: 10.3109/19401736.2015.1074201. [DOI] [PubMed] [Google Scholar]
- Matsumoto R. 2001. Shiranuhi: a late-maturing citrus cultivar. Bull Natl Inst Fruit Tree Sci. 35:115–120. [Google Scholar]
- Penjor T, Yamamoto M, Uehara M, Ide M, Matsumoto N, Matsumoto R, Nagano Y.. 2013. Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS One 8:e62574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HS, Park YH, Moon DG.. 2005. Volatile flavor properties of Hallabong grown in open field and green house by GC/GC-MS and sensory evaluation. J Korean Soc Food Sci Nutr. 34:1239–1245. [Google Scholar]
- Su HJ, Hogenhout SA, Al-Sadi AM, Kuo CH.. 2014. Complete chloroplast genome sequence of Omani lime (Citrus aurantiifolia) and comparative analysis within the rosids. PLoS One 9:e113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F, Perrier X, Ruiz M, Scalabrin S, Terol J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 32:656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, et al. 2013. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 45:59–66. [DOI] [PubMed] [Google Scholar]
- Zhang D, Mabberley DJ.. 2008. Citrus. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 11. Beijing: Science Press/St. Louis, MO: Botanical Garden Press; [cited 2015 Nov 18]. Available from: http://efloras.org/florataxon.aspx?flora_id=2&taxon_id=107164. [Google Scholar]