Abstract
The complete mitochondrial genome of the zebra shark (Stegostoma fasciatum) was first determined in this study. The total length of this circle DNA was 16 658 bp, consisted of 37 genes with typical gene order in vertebrate mitogenome. Its nucleotide content was 34.1% A, 25.9% C, 12.5% G and 27.5% T. This mitogenome had 23 bp short intergenic spaces located in 10 gene junctions and 54 bp overlaps located in 11 gene junctions. In the protein-coding genes, two start codons (GTG and ATG) and two stop codons (TAG and TAA/T) were found. The 22 tRNA genes ranged from 66 bp (tRNA-Cys) to 75 bp (tRNA-Leu1). The phylogenetic result showed that S. fasciatum was clustered with the whale shark Rhincodon typus.
Keywords: Orectolobiformes, Stegostoma fasciatum, Stegostomatidae
The zebra shark (Stegostoma fasciatum) (Stegostomatidae, Orectolobiformes) is found on sand, rubble or coral bottoms of the continental and insular shelves in the tropic, of Indo-West Pacific Oceans (Compagno 1984; Randall et al. 2003). The molecular and genetic research of this species was rare. In this study, we first determined the complete mitochondrial genome of S. fasciatum, and analyzed the phylogenetic relationship of the sharks in Orectolobiformes fishes.
Tissue of S. fasciatum (voucher no. RN2012122333) was collected from Ranong, Thailand. The experimental protocol and data analysis methods followed Chen et al. (2014). Excluding S. fasciatum and the outgroup Chimaera monstrosa, five species of Orectolobiformes, as well as each two species of Carcharhiniformes, Heterodontifomes and Lamniformes, with the complete mitogenomes available in the Genbank, were selected to construct the phylogenetic tree. The Bayesian method was fulfilled with the GTR + I+G model by three partitions of the mitogenome: 12S and 16S rRNA genes, and the first and second codons of the 12 protein-coding genes (except the light-strand encoded ND6 gene).
The total length of the complete mitochondrial DNA of S. fasciatum was 16 658 bp (Genbank accession no. KU057952). Its nucleotide base composition was 34.1% A, 25.9% C, 12.5% G and 27.5% T. This circle molecular had a typical mitogenomic organization and gene order as most vertebrates, containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one non-coding control region. Whole mitogenome had 23 bp short intergenic spaces located in 10 gene junctions and 54 bp overlaps located in 11 gene junctions. The 13 protein-coding genes used two start codons (GTG and ATG) as well as two stop codons (TAG and TAA/T), and most of them shared common initial codon ATG and terminal codon TAA/T. The COI gene owned a non-standard initial codon GTG, which is common in vertebrates (Slack et al. 2003). The COII, ND4 and Cytb genes were terminated with a single T, whereas the ND1, ND3 and ND6 genes were terminated with the TAG. The 22 tRNA genes ranged from 66 bp (tRNA-Cys) to 75 bp (tRNA-Leu1), among which six tRNA genes were located in the light strand, while the remaining tRNA genes were in the heavy strand. The 12S and 16S rRNA genes were located between the tRNA-Phe and tRNA-Leu1 genes, separated by the tRNA-Val gene. A 38 bp inserted sequence was identified as the origin of light-strand replication (OL) between tRNA-Asn and tRNA-Cys genes with a stem-loop structure. The control region was 1000 bp, presenting a high A + T content (69.3%).
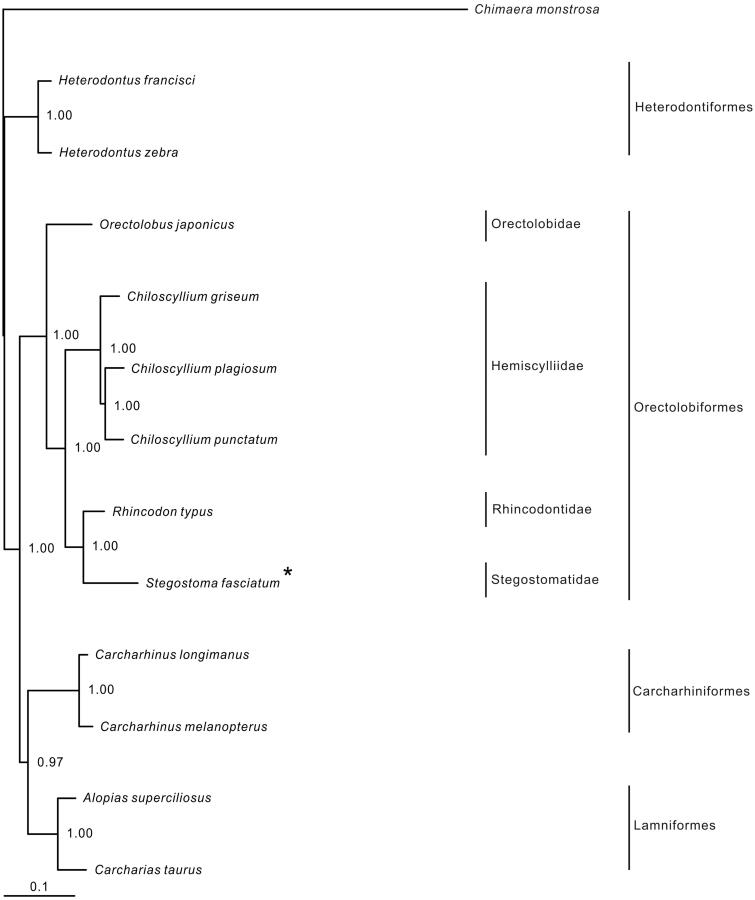
In the Bayesian tree, all nodes were strongly supported (Figure 1). Each order was monophyletic. The Charcharhiniformes was the sister to the Lamniformes. The Orectolobiformes was clustered to the (Charcharhiniformes + Lamniformes) clade, and the Heterodontiformes was the basal clade in the tree. Within the Orectolobiformes, four available families formed a (Orectolobidae + (Hemiscylliidae + (Rincodontidae + Stegostomatidae))) relationship. Stegostoma fasciatum was clustered with the whale shark Rhincodon typus.
Figure 1.
Phylogenetic position of Stegostoma fasciatum. Chimaera monstrosa (AJ310140.1) was selected as the outgroup. The six species from the order Orectolobiformes were Stegostoma fasciatum (KU057952), Chiloscyllium griseum (JQ434458), C. plagiosum (NC_012570.1), C. punctatum (JQ082337.1), Orectolobus japonicus (KF111729.1) and Rhincodon typus (KC633221.1). The two species from Carcharhiniformes were Carcharhinus longimanus (NC_025520.1) and C. melanopterus (NC_024284.1). The two species from Heterodontifomes were Heterodontus francisci (NC_003137.1) and H. zebra (KC845548.1). The two from Lamniformes were Alopias superciliosus (KC757415.1) and Carcharias taurus (KF569943.1).
Declaration of interest
This study was supported by the Ministry of Science and Technology of Zhejiang Province (2013F50015), the Ministry of Science and Technology of Wenzhou City (S20140013) and Guangdong Natural Science Foundation (2015A030310002). The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Chen X, Ai W, Xiang D, Chen S.. 2014. Complete mitochondrial genome of the red stingray Dasyatis akajei (Myliobatiformes: Dasyatidae). Mitochondrial DNA. 25:37–38. [DOI] [PubMed] [Google Scholar]
- Compagno LJV. 1984. FAO species catalogue. Vol. 4. Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 1 – Hexanchiformes to Lamniformes. FAO Fish. Synop, 125(4/1). Rome: FAO. p. 1–249. [Google Scholar]
- Randall JE, Williams JT, Smith DG, Kulbicki M, Tham GM, Labrosse P, Kronen M, Clua E, Mann BS.. 2003. Checklist of the shore and epipelagic fishes of Tonga. Atoll Res Bull Nos 502:12. [Google Scholar]
- Slack KE, Janke A, Penny D, Arnason U.. 2003. Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene 302:43–52. [DOI] [PubMed] [Google Scholar]