Abstract
In this study, the complete mitochondrial genome of Marco Polo wild sheep was sequenced for the first time. It is 16 613 bp in length and possesses 22 tRNA genes, 13 typical mammalian protein-coding genes, two rRNA genes and one D-loop region. The whole genome’s base composition is 33.7% A, 25.8% C, 13.1% G and 27.4% T, and the percentage of AT-rich regions is 61.1%.
Keywords: Endangered species, evolution, mitochondrial genome, wild sheep
Argali (Ovis ammon) are the largest wild sheep in the world and are threatened throughout their habitat in central Asia (Kenny et al. 2008; IUCN 2013). Currently, nine different subspecies are commonly recognized (Fedosenko & Blank 2005; Wilson & Reeder 2005), of which, Marco Polo Argali (Ovis ammon polii/O. a. polii) have the longest horns. Due to illegal and legal hunting, habitat loss and competition with livestock (Reading et al. 2001), they are now facing an unprecedented threat throughout their habitat (geospatial coordinates: 37°05∼39°30′ N-71°0′∼75°50′ E), which is adjacent to the borders of China, Pakistan, Afghanistan and Tajikistan (Schuerholz 2001). This study is the first published mitochondrial genome of O. a. polii (GenBank accession number: KT781689), which can be used to investigate evolutionary relationships among Ovis species and the Argali taxonomy.
Mitochondrial DNA from one specimen of O. a. polii blood was collected form Tianshan Mountain Wildlife Park (Urumqi, Chian) and sequenced using Illumina Hiseq 2500 (Illumina Inc., San Diego, CA). The mitogenome was successfully assembled using six scaffolds and six gap-filling polymerase chain reaction (PCR) products from Sanger sequencing. It is 16 613 bp in length, and harbours 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a non-coding D-Loop RNA. Its overall elements, and the gene arrangement pattern, are similar to those of the typical vertebrate mitogenome. The overall base composition (A: 33.7%, C: 25.8%, G: 13.1% and T: 27.4%) results in a low G + C (38.9%) content. Eight tRNA were encoded on the light strand, with the remainder of the genes encoded on the heavy strand. Of the 13 coding proteins, 10 use ATG as the start codon and three use ATA. Additionally, seven open-reading frames end with TAA, three genes use T, and single gene uses TAG, AGA and TA as stop codons. The lengths of the 12s and 16s RNA genes located between tRNAphe and tRNAleu are 958 and 1575 bp, respectively.
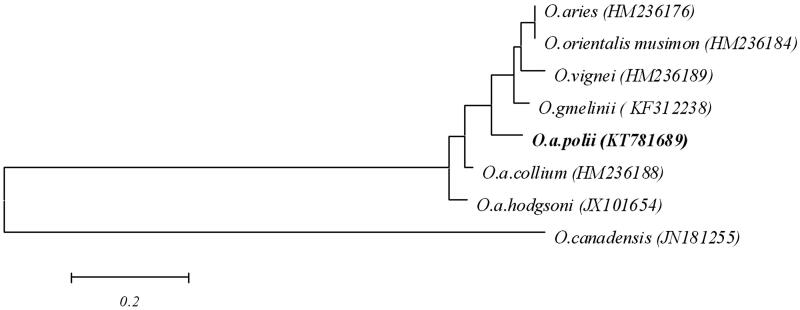
The complete O. a. polii mitogenome was compared with five Ovis species [O. vignei (HM236189), O. canadensis (JN181255), O. orientalis musimon (HM236184), O. gmelinii (KF312238) and O. aries (HM236176)] and two argali subspecies[O. a. collium (HM236188 and O. a. hodgsoni (JX10165)] (Hiendleder et al. 1998; Lancioni et al. 2013; Hu et al. 2016). The gene order and base composition were conserved among these species, but the D-loop-region’s sequences belong to a highly varied region among the species. The phylogenetic analysis indicated that the whole mitogenome of O. a. polii showed a high level of conservation with three wild species (Figure 1), O. vignei, O. gmelinii, O. ammon (O. a. collium and O. a. hodgsoni), as compared with domesticated sheep (O. aries) and bighorn wild sheep (O. Canadensis).
Figure 1.
Neighbor-joining tree for five wild Ovis species and domesticated sheep based on the sequence of whole mitogenome.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by grants from the Hi-Tech Research and Development Program of China (863) (No. 2012AA021801), the National Natural Science Foundation of China (Nos. 31260281 and 31160452).
References
- Fedosenko AK, Blank DA.. 2005. Mammalian Species, Ovis ammon. Am Soc Mammal. 773:1–15. [Google Scholar]
- Hiendleder S, Lewalski H, Wassmuth R, Janke A.. 1998. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J Mol Evol. 47:441–448. [DOI] [PubMed] [Google Scholar]
- Hu X, Gao L.. 2016. The complete mitochondrial genome of domestic sheep, Ovis aries. Mitochondrial DNA 27:1425–1427. [DOI] [PubMed] [Google Scholar]
- IUCN 2013. IUCN Red List of Threatened Species. Version 2013.1. Available from: www.iucnredlist.org.
- Kenny D, Denicola A, Amgalanbaatar S, Namshir Z, Wingard G, Reading R.. 2008. Successful field capture techniques for free-ranging argali sheep (Ovis ammon) in Mongolia. Zoo Biol. 27:137–144. [DOI] [PubMed] [Google Scholar]
- Lancioni H, Di Lorenzo P, Ceccobelli S, Perego UA, Miglio A, Landi V, Antognoni MT, Sarti FM, Lansagna E, Achilli A.. 2013. Phylogenetic relationships of three Italian merino-derived sheep breeds evaluated through a complete mitogenome analysis. PLoS One 8:e73712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading RP, Amgalanbaatar S, Wingard G.. 2001. Argali sheep conservation and research activities in Mongolia. The Open Country 3:25–32. [Google Scholar]
- Schuerholz G. 2001. Community based wildlife management (CBWM) in the Altai Sayan Ecoregion of Mongolia feasibility assessment: opportunities for and barriers to CBWM. Ulaanbaatar: WWF-Mongolia; . [Google Scholar]
- Wilson DE, Reeder DM.. 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd ed Baltimore: The Johns Hopkins University Press. [Google Scholar]