Abstract
The complete mitochondrial genome of the Greek nine-spined stickleback Pungitius hellenicus was obtained using Illumina high-throughput sequencing of genomic DNA. The genome was 16 713 bp long, and contained 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a control region. The arrangement of the genes was identical to that of other Gasterosteidae fishes. However, the control region of P. hellenicus contained three copies of imperfect repeated sequences (72–78 bp in single motifs), while P. pungitius and P. tymensis have one or two copies. Nucleotide identity between P. hellenicus and three other Pungitius species across all the 37 genic regions was 93.0% to 95.5%.
Keywords: Gasterosteidae, genome, mtDNA, Pungitius hellenicus
The number of valid species in the circumpolarly distributed teleost fish genus Pungitius remains uncertain, but morphological evidence suggests that the Greek nine-spined stickleback (Pungitius hellenicus) is likely to be a valid species (Keivany et al. 1997; Keivany & Nelson 2000). However, little genetic information is available on this species (Geiger et al. 2014). Hence, access to the complete mitochondrial genome sequence of P. hellenicus would allow evaluation of its validity as a distinct species, as well as its phylogenetic positioning within the genus Pungitius (cf., Takahashi & Goto 2001; Keivany & Nelson 2004; Mattern 2004). As the known distribution of P. hellenicus is confined to only a few localities in the Sperchios River basin in Central Greece (Keivany et al. 1999; Keivany & Nelson 2000), and the species is listed as critically endangered (Martins & Wiswedel 2015), the genomic information is likely to support conservation and management planning of this species.
Pungitius hellenicus was collected from the Sperchios River basin in Greece (38°51′N, 22°26′E), and genomic DNA was sequenced using the Illumina HiSeq2000 platform with 100 paired-end strategy. In total, 16.6 million reads were aligned against the reference P. sinensis mitogenome (Hwang et al. 2012a) using bwa-0.5.10 (Li & Durbin 2009). Mean sequence coverage across the mitogenome was 43-fold, and with the exception of a 265 bp gap in the control region, all regions had at least one-fold coverage (83.7% with ≥20-fold coverage). Sanger sequencing was used to fill the 265 bp gap. PCR products were obtained using a primer set (PCR-F: 5′-CAAGGTTGAACATCTTTCGC-3′ and PCR-R: 5′-CTGATACCAGCTCCTTGTTCC-3′) following the protocols and procedures described in Teacher et al. (2011). The consensus sequence of the P. hellenicus mitogenome was generated using SAMtools 1.2 (Li et al. 2009) and manually checked.
The complete mitochondrial genome of P. hellenicus was 16 713 bp long (GenBank Accession No. KU236383), containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a control region. The arrangement of the genes was identical to that of other Gasterosteidae fishes (Miya et al. 2001; Kawahara et al. 2009; Hwang et al. 2012a, 2012b). In the 13 protein-coding genes, an incomplete stop codon was found for four genes (ND2, COII, ND4 and Cytb). Three copies of imperfect repeated sequences (72–78 bp in single motifs) were identified in the control region, whereas one or two copies have been observed in P. pungitius and P. tymensis (Takahashi & Goto 2001). The overall base composition of the entire mitogenome was 27.4% for A, 26.4% for T, 17.6% for G and 28.6% for C.
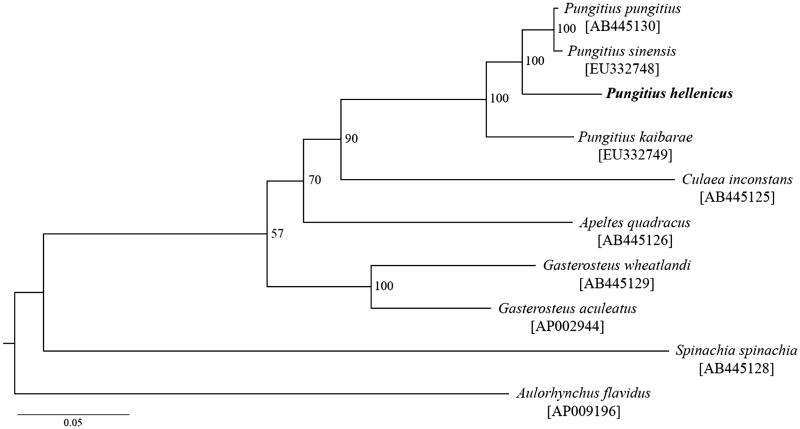
To investigate the phylogenetic position of P. hellenicus in Gasterosteidae fishes, a maximum-likelihood tree was constructed with the 37 genes (15 581 bp in total) using RAxML v.8.0 under the GTR + GAMMA model, 37 gene partitions and 100 thorough bootstrap replicates (Stamatakis 2014). Pungitius hellenicus was phylogenetically positioned with other Pungitius species (i.e. P. pungitius, P. sinensis and P. kaibarae), showing clear divergence from them (Figure 1). Nucleotide identity in the 37 gene regions was 93.0% to 95.5% between P. hellenicus and the other Pungitius species.
Figure 1.
A maximum likelihood tree inferred from 37 mitochondrial genes among nine Gasterosteidae and an outgroup (Aulorhynchus flavidus) species. Bootstrap support is indicated at nodes. GenBank accession numbers are indicated in brackets.
Acknowledgements
We thank Pekka Ellonen, Laura Häkkinen, Tiina Hannunen and Sami Karja for help with laboratory and bioinformatic work. The sequencing was conducted at Finnish Institute for Molecular Medicine.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by grants (108601 & 118673) from the Academy of Finland, the Ministry of Culture of the Czech Republic (DKRVO 2015/15, National Museum, 00023272) and by the institutional resources of the Ministry of Education, Youth and Sports of the Czech Republic.
References
- Geiger MF, Herder F, Monaghan MT, Almada V, Barbieri R, Bariche M, Berrebi P, Bohlen J, Casal-Lopez M, Delmastro GB, et al. 2014. Spatial heterogeneity in the Mediterranean Biodiversity Hotspot affects barcoding accuracy of its freshwater fishes. Mol Ecol Resour. 14:1210–1221. [DOI] [PubMed] [Google Scholar]
- Hwang D-S, Song HB, Lee J-S.. 2012a. Complete mitochondrial genome of the Amur stickleback Pungitius sinensis (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:293–294. [DOI] [PubMed] [Google Scholar]
- Hwang D-S, Song HB, Lee J-S.. 2012b. Complete mitochondrial genome of the Amur stickleback Pungitius kaibarae (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:313–314. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M.. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50:401–404. [DOI] [PubMed] [Google Scholar]
- Keivany Y, Nelson JS, Economidis PS.. 1997. Validity of Pungitius hellenicus Stephanidis, 1971 (Teleostei, Gasterosteidae), a stickleback fish from Greece. Copeia 1997:558–564. [Google Scholar]
- Keivany Y, Daoulas CK, Nelson JS, Economidis PS.. 1999. Threatened fishes of the world: Pungitius hellenicus Stephanidis, 1971 (Gasterosteidae). Environ Biol Fish. 55:390–390. [Google Scholar]
- Keivany Y, Nelson JS.. 2000. Taxonomic review of the genus Pungitius, ninespine sticklebacks (Gasterosteidae). Cybium 24:107–122. [Google Scholar]
- Keivany Y, Nelson JS.. 2004. Phylogenetic relationships of sticklebacks (Gasterosteidae), with emphasis on ninespine sticklebacks (Pungitius spp.). Behaviour 141:11–12. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins J, Wiswedel S.. 2015. Pungitius hellenicus The IUCN Red List of Threatened Species 2015: e.T18875A19928983 [cited 2015 Dec 5]. Available from: 10.2305/IUCN.UK.2015-2.RLTS.T18875A19928983.en. [DOI] [Google Scholar]
- Mattern MY. 2004. Molecular phylogeny of the Gasterosteidae: the importance of using multiple genes. Mol Phylogenet Evol. 30:366–377. [DOI] [PubMed] [Google Scholar]
- Miya M, Kawaguchi A, Nishida M.. 2001. Mitogenetic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Phylogenet Evol. 18:1993–2009. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Goto A.. 2001. Evolution of East Asian ninespine sticklebacks as shown by mitochondrial DNA control region sequences. Mol Phylogenet Evol. 21:135–155. [DOI] [PubMed] [Google Scholar]
- Teacher AG, Shikano T, Karjalainen ME, Merilä J.. 2011. Phylogeography and genetic structuring of European nine-spined sticklebacks (Pungitius pungitius)-mitochondrial DNA evidence. PLoS ONE 6:e19476. [DOI] [PMC free article] [PubMed] [Google Scholar]