Abstract
Known colloquially as ‘Old Man’s Beard’, Usnea is a genus of lichenized Ascomycete fungi characterized by having a fruticose growth form and cartilaginous central axis. The complete mitochondrial genomes of Usnea halei, U. mutabilis, U. subfusca, U. subgracilis, and U. subscabrosa were sequenced using Illumina data and then assembled de novo. These mitogenomes ranged in size from 52,486 bp (U. subfusca) to 94,464 bp (U. subgracilis). All were characterized by having high levels of intronic and intergenic variation, such as ORFs that encode proteins with homology to two homing endonuclease types, LAGLIDADG and GIY-YIG. Genes annotated within these mitogenomes include 14 protein-coding genes, the large and small ribosomal subunits (LSU and SSU), and 23–26 tRNAs. Notably, the atp9 gene was absent from each genome. Genomic synteny was highly conserved across the five species. Five conserved mitochondrial genes (nad2, nad4, cox1, cox2, and cox3) were used to infer a best estimate maximum likelihood phylogeny among these five Usnea and other relatives, which yielded relationships consistent with prior published phylogenies.
Keywords: Symbiosis, Usnea halei, Usnea mutabilis, Usnea subfusca, Usnea subgracilis, Usnea subscabrosa
Introduction
Usnea is a genus of lichenized Ascomycete fungi that represents one of the most species-rich genera in Parmeliaceae, which is a family of more than 2000 species and 77 genera (Crespo et al. 2007; Lücking et al. 2016). Usnea has a cosmopolitan distribution, being found on every continent (Halonen 2000), and contains both saxicolous (rock-dwelling) and corticolous (bark-dwelling) species (Clerc and Herrera-Campos 1997; Clerc 1998; Halonen 2000). The genus on the whole is characterized by having a fruticose growth form and a cartilaginous central axis (Clerc 1998). Known colloquially as ‘Old Man’s Beard’, species of Usnea are charismatic and highly recognizable lichens across the globe (Brodo et al. 2001). Species are characteristically yellowish-green in colour due to the production of usnic acid, which is thought to have a multitude of potential functions including having antibiotic, antimycotic, and antiprotozoal properties (Lauterwein et al. 1995; Fournet et al. 1997; Giordano et al. 1997; Cocchietto et al. 2002). Here, we present newly sequenced and annotated mitochondrial genomes from five species of Usnea that occur in the southern Appalachian lichen biodiversity hotspot, located in eastern North America (Lendemer et al. 2013; Tripp and Lendemer in press; Tripp et al. in review). Two of these species (U. halei and U. subfusca) are endemic to eastern North America and have centres of distributions in the region (Clerc and Herrera-Campos 1997; Lendemer et al. 2013).
Methods
Samples of Usnea halei (voucher Lendemer 46374; Lat/Long: 35.1356,–83.1911; NCBI accession MG722979), U. mutabilis (voucher Lendemer 49260; Lat/long: 34.3679,–85.6299; NCBI accession MG920803), U. subfusca (voucher Lendemer 46309; Lat/Long: 35.61,–83.4467; NCBI accession MG720812), U. subgracilis (voucher Lendemer 48717; Lat/Long: 35.1036,–83.2075; NCBI accession MG720066), and U. subscabrosa (voucher Lendemer 46747; Lat/long: 35.4372,–83.7481; NCBI accession MG720452) were collected by J.C. Lendemer, E.A. Tripp, K.G. Keepers and K.H. White from the Southern Appalachian Mountains in the United States between 2015 and 2017. Genomic DNA was extracted using a Qiagen DNeasy 96 plant kit, with the protocol modified to include a 10 min 65 °C incubation step for ground material in lysis buffer and a 100% ethanol wash before final drying of the membrane prior to elution. Genomic libraries were prepared using Nextera® XT DNA library prep kits (Illumina®) and each sample was barcoded using the unique dual index adapters Nextera® i5 and i7. Samples that passed QC were processed for paired-end 150 base pair read sequencing on the Illumina NextSeq® at University of Colorado’s BioFrontiers Institute Next-Generation Sequencing Facility in Boulder, Colorado. Genomic reads were trimmed using Trimmomatic-0.36 with the parameters ‘ILLUMINACLIP:NexteraPE-PE.fa:2:20:10 MINLEN:140 LEADING:20 TRAILING:20’ (Bolger et al. 2014). Trimmed reads were assembled de novo using SPAdes v.3.9, with the parameters ‘-meta -k 35,55,85’ (Bankevich et al. 2012). Fungal mitochondrial contigs were identified using a command-line BLAST against a set of fungal mitochondrial proteins derived from the lichenized fungus Peltigera dolichorrhiza (NCBI accession KT946595), which were then circularized and error-corrected. Annotations of genomic features were initiated using DOGMA (Wyman et al. 2004) and completed in NCBI’s Sequin 15.10 (Bethesda, MD). Genome assemblies and annotations were performed by undergraduate and graduate students enrolled in N. Kane's Genomics class at the University of Colorado, Boulder.
Phylogenetic analysis
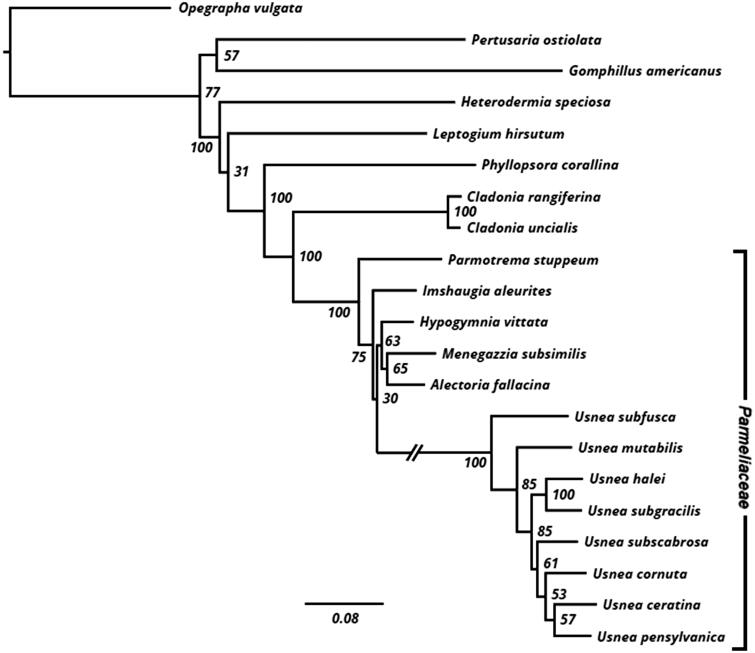
A maximum likelihood phylogenetic hypothesis was constructed using sequence data from five loci (cox3, nad2, nad4, cox1, and cox2), which were concatenated prior to analysis. Outgroup sequences were obtained from NCBI: O. vulgata [KY315997], P. ostiolata [KY346830], G. americanus [NC_034790], H. speciosa [KY328643], L. hirsutum [NC_034928], P. corallina [NC_034779], C. rangiferina [KY460674], C. uncialis [KY352404], P. stuppeum [KY362439], I. aleurites [KY352227], H. vittata [KY362374], M. subsimilis [KY352491], A. fallacina [MG711470], U. cornuta [KY100278], U. ceratina [KX987159], and U. pensylvanica [KY321923]. Sequences were aligned in MEGA v.7.0.26 (Kumar et al. 2016) using the MUSCLE aligner (Edgar 2004). The resulting alignment was manually manipulated to exclude regions of the alignment that contained excessive missing data (available at Dryad Zenodo, record 1185346). Gaps were treated as missing data. The ML tree was inferred using RAxML v.8.2.10 (Stamatakis 2014). To assess support among relationships, we conducted 250 bootstrap replicates using the autoMRE criterion in RAxML and from this distribution, a consensus topology was generated. Bootstrap support values were mapped onto the most likely tree deriving from the above search. The tree was rooted using Opegrapha vulgata.
Results and discussion
Mitochondrial genome content and organization
The lengths of Usnea mitochondrial genomes in this study varied from 52,486 bp (U. subfusca) to 94,464 bp (U. subgracilis). The mitochondrial genomes of the other three species were of intermediate lengths: 61,314 bp (U. mutabilis), 78,464 bp (U. subscabrosa), and 82,851 bp (U. halei). The genomes presented here contained a conserved set of 14 protein coding genes (cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, atp6, atp8, and rps3). Notably, all lacked a key mitochondrial gene (atp9), which has previously been shown to have been lost evolutionarily from three different lineages of lichens, one of which includes Parmeliaceae (Pogoda et al. 2018). Synteny of protein-coding genes in the mitogenomes was highly conserved across the five species of Usnea, with no differences in overall gene order and no inversions detected.
Introns and intergenic spacer regions
Among the five samples, non-coding DNA contributed to overall genome size: U. subfusca has the smallest mitochondrial genome and also the least amount of intronic sequence. The three largest genomes (U. subscabrosa, U. halei, and U. subgracilis) contained variable lengths of intronic sequences, but all had approximately two to four times more total intergenic sequence compared to the other genomes. Notably, the three largest genomes also contained a large intergenic region between atp8 and cob, which contained multiple fragments of a DNA polymerase (dpo) and ranged in size from approximately 4 kb in U. subgracilis to 17.9 kb in U. subscabrosa. All five genomes contained a large intergenic gap between the LSU and nad2 genes, ranging in size from 4.8 to 7.7 kb and containing 7–13 tRNA sequences. Usnea subscabrosa also contained an ORF with homology to a GIY-YIG homing endonuclease in this region.
Open reading frames
Usnea mutabilis had multiple ORFs throughout its mitogenome. The longest ORF (1193 bp) was found preceding nad5 and is likely co-transcribed with a 3′ ORF that shares the stop codon of the protein coding gene. Two hypothetical proteins were located between nad4 and rps3 and between cox1 and nad4, having lengths of 311 bp and 758 bp, respectively. We found that U. mutabilis possessed three ORFs with homology to homing endonucleases located in the introns of the cox1 and LSU genes. Usnea subfusca was found to have two ORFs, one with homology to a GIY-YIG homing endonuclease that was 1445 bp and was located at the 3′ end of the nad1 gene.
Phylogenetic history
All eight species of Usnea (the five presented here, along with three previously sequenced species obtained from NCBI) were recovered as monophyletic with 100% bootstrap support (Figure 1). Monophyly of Parmeliaceae as a whole was also well supported (BS = 100%), with Imshaugia, Hypogymnia, Menegazzia, and Alectoria recovered as sister to Usnea. This topology is in general agreement with previously published phylogenies (e.g. Crespo et al. 2010; Miadlikowska et al. 2014).
Figure 1.
Phylogenetic relationships among Usnea and other lichens inferred using maximum likelihood (see Methods section). Numbers next to nodes represent bootstrap support mapped onto the most likely tree (–ln L = 68803.11). The long branch leading to Usnea was shortened (hashes) for the figure.
Conclusions
The five newly sequenced, assembled, and annotated Usnea mitogenomes exhibited a large range in size, with intronic and intergenic regions of variable lengths associated with changes in genome size. These mitochondrial genomes coded for a variable set of ORFs, but all contained the same set of 14 protein-coding genes. Results from our study confirm earlier findings (Pogoda et al. 2018) that members of Parmeliaceae have lost a key gene involved in energy production, atp9, lending genomic evidence of the obligate nature of this symbiotic relationship. We thus infer that these five species of Usnea are likely dependent on their photosynthetic partner(s) for energy production. Finally, these data provide an important resource for further study of the evolution of parasitic, selfish elements such as LAGLIDADG and GIY-YIG type homing endonucleases in Usnea and related lichens. The present study yields new resources for further study of lichen mitochondrial genome evolution and co-evolution with obligate symbiotic partners.
Acknowledgements
Special thanks to the students of University of Colorado Boulder’s Genomics classes of 2016 and 2017 for assembling and annotating the genomes used in this study. Publication of this chapter was funded by the University of Colorado Boulder Libraries Open Access Fund.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content of this paper.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD.. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30;2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodo IM, Sharnoff SD, Sharnoff S.. 2001. Lichens of North America. Conn, New Haven: Yale University Press. [Google Scholar]
- Clerc P, Herrera-Campos MA.. 1997. Saxicolous species of Usnea subgenus Usnea (lichenized ascomycetes) in North America. Bryologist. 281–301. [Google Scholar]
- Clerc P. 1998. Species concepts in the genus Usnea (lichenized Ascomycetes). Lichenologist. 30:321–340. [Google Scholar]
- Cocchietto M, Skert N, Nimis P, Sava G.. 2002. A review on usnic acid, an interesting natural compound. Naturwissenschaften. 89:137–146. [DOI] [PubMed] [Google Scholar]
- Crespo A, Kauff F, Divakar PK, del Prado R, Pérez-Ortega S, de Paz GA, Ferencova Z, Blanco O, Roca-Valiente B, Núñez-Zapata J, et al. . 2010. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon. 59:1735–1753. [Google Scholar]
- Crespo A, Lumbsch HT, Mattsson JE, Blanco O, Divakar PK, Articus K, Wiklund E, Bawingan PA, Wedin M.. 2007. Testing morphology-based hypotheses of phylogenetic relationships in Parmeliaceae (Ascomycota) using three ribosomal markers and the nuclear RPB1 gene. Mol Phylogenet Evol. 44:812–824. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournet A, Ferreira ME, de Arias AR, de Ortiz ST, Inchausti A, Yalaff G, Quilhot W, Fernandez E, Hidalgo ME.. 1997. Activity of compounds isolated from Chilean lichens against experimental cutaneous leishmaniasis. Comp Biochem Physiol C: Pharmacol Toxicol Endocrinol. 116:51–54. [DOI] [PubMed] [Google Scholar]
- Giordano S, Basile A, Lanzetta R, Corsaro MM, Spagnuolo V, Castaldo Cobianchi R.. 1997. Potential allelochemicals from the lichen Cladonia foliacea and their in vitro effects on the development of mosses. Allelopathy J. 4:89–100. [Google Scholar]
- Halonen P. 2000. Studies on the lichen genus Usnea in East Fennoscandia and Pacific North America (PDF). Oulu, Finland: Oulu University Library; p. 13. ISBN 9514255232. ISSN 0355-3191. [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterwein M, Oethinger M, Belsner K, Peters T, Marre R.. 1995. In vitro activities of the lichen secondary metabolites vulpinic acid, (+)-usnic acid, and (–)-usnic acid against aerobic and anaerobic microorganisms. Antimicrob Agents Chemother. 39:2541–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendemer JC, Harris RC, Tripp EA.. 2013. The lichens and allied fungi of Great Smoky Mountains National Park. Memoires NY Bot Gard. 104:1–152. [Google Scholar]
- Lücking R, Hodkinson BP, Leavitt SD.. 2016. The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota – approaching one thousand genera. Bryologist. 119:361–416. [Google Scholar]
- Miadlikowska J, Kauff F, Högnabba F, Oliver JC, Molnár K, Fraker E, Gaya E, Hafellner J, Hofstetter V, Gueidan C, et al. . 2014. A multigene phylogenetic synthesis for the class Lecanoromycetes (Ascomycota): 1307 fungi representing 1139 infrageneric taxa, 317 genera and 66 families. Mol Phylogenet Evol. 79:132–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda CS, Keepers KG, Lendemer JC, Kane NC, Tripp EA.. 2018. Reductions in complexity of mitochondrial genomes in Lichen-Forming fungi shed light on genome architecture of obligate symbioses. Mol Ecol. doi: 10.1111/mec.14519. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp E, Lendemer JC, McCain CM.. in review. Impacts of disturbance on lichens – obligate symbiotic organisms—in a temperate biodiversity hotspot. Divers Distrib. [Google Scholar]
- Tripp EA, Lendemer JC.. in press. Field guide to the Lichens of Great Smoky Mountains National Park. Knoxville (TN): University of Tennessee Press. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]