Abstract
Chloroplasts (cp) are indispensable organelles in plant cells that perform photosynthesis amongst other functions, including producing pigments, synthesizing sugars and certain amino acids. The complete chloroplast genome of Tartary Buckwheat Cultivar Miqiao 1 is sequenced in this study. Miqiao 1 is currently the only Tartary Buckwheat variety that can produce the whole nutritive Tartary Buckwheat Rice by hulling. The Miqiao 1 cp genome size is 159,272 bp in length, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. A pair of inverted repeats (IRs) of 30,817 bp were disconnected by a large single copy (LSC) of 84,397 bp and a small single copy (SSC) of 13,241 bp. The cp genome with 37.9% GC content contained 113 annotated genes.
Keywords: Complete chloroplast genome, medicinal plant, Tartary Buckwheat Cultivar Miqiao 1
Chloroplasts (cps) are indispensable plant cell organelles that conduct photosynthesis in the presence of daylight. The chloroplast genome in land plants is a circular molecule ranging in size from 76 to 217 kb, with a highly conserved structure of two copies of large inverted repeats (IR) detached by small (SSC) and large (LSC) single-copy regions (Dane et al. 2015). Characteristics of the cp genome have been used comprehensively as a tool to investigate evolutionary relationships and molecular phylogenetic of seed plants (Jansen et al. 2011).
Buckwheat (Fagopyrum species) which belongs to Polygonaceae, a member of knotgrass, is an annual herbaceous plant. Buckwheat is classified into 20 species. Geographically, it is principally centred in the Eurasian region and grown in the highlands. Further, it is an extensively cultivated multipurpose crop (Kump & Javornik 1996; Ohsako et al. 2002). Tartary buckwheat is an exceptionally opulent source of rutin compared to common buckwheat. Rutin assists moderate diabetes, high blood pressure and intravascular cholesterol (Fabjan et al. 2003). It is also reported to have a decisive role in pharmaceutical research (Kim & Kim 2004). Miqiao 1 is currently the Tartary Buckwheat variety that can produce the Whole Nutritive Tartary Buckwheat Rice by hulling. Recently, the cp genome sequence of some plants have been reported, but medicinal plants are rarely reported (Dane et al. 2015). Three species of Fagopyrum Mill have been reported recently (Logacheva et al. 2008; Cho et al. 2015; Yang et al. 2015).
The genomic DNA was extracted from the leaves of Fagopyrum tataricum Gaertn. cultivated in the field of College of Life Science, Sichuan Agricultural University, Ya’an, China. This study reports the sequencing of the Miqiao 1. The whole cp genome was sequenced using HiSeq 4000 PE150 Illumina (Biocode Biotech Co., Beijing, China) and assembled into the complete cp genome using Sequencher version 4.10 (Bankevich et al. 2012). DualOrganellarGenoMe Annotator (Wyman et al. 2004) was used to annotate the cp genome with BLASTX and BLASTN identifying the location of encoding genes and RNA (Altschul et al. 1997). The complete genome sequence was submitted to GenBank under the accession number KX085498.
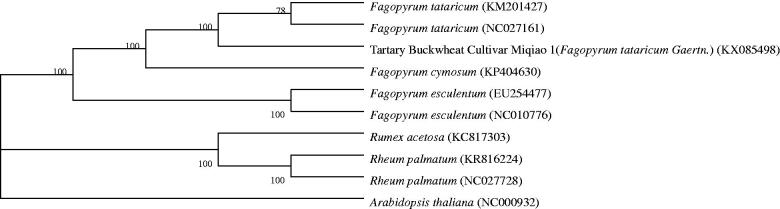
The total size of complete cp genome was 159,272 bp with 37.9% GC content, consisting of one LSC (84,397 bp), one SSC (13,241 bp) and two IRs (30,817 bp). The GC content of the IRs (41.3%) was higher than that of LSC and SSC (36.2% and 32.8%, respectively). A total of 113 genes were successfully annotated, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. There were 17 intron-containing genes, all of which were single-intron genes except for ycf3 and clpP with two introns separately. rpsl2 was a reverse-splicing gene, of which the 5′ end exon located in the LSC region and 3′ end exon was in the IR region. The phylogenetic relationship of the Miqiao 1 was deduced by comparing it with other nine chloroplast genomes downloaded from the GenBank (Figure 1).
Figure 1.
ML tree based on Tartary Buckwheat Cultivar Miqiao 1 chloroplast genomic sequence compared with 9 sequences obtained from NCBI.
Acknowledgments
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This work is supported by funding from the Department of Science and Technology of Sichuan Province (No. 2014HH0012).
References
- Altschul SF, Madden TL, Schffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD.. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing . J Comput Biol 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KS, Yun BK, Yoon YH, Hong SY, Mekapogu M, Kim KH, Yang TJ.. 2015. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One. 10:e0125332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane F, Wang Z, Goertzen L.. 2015. Analysis of the complete chloroplast genome of Castanea pumila var. pumila, the Allegheny chinkapin. Tree Genet Genomes. 11:1–6. [Google Scholar]
- Fabjan N, Rode J, Košir IJ, Wang Z, Zhang Z, Kreft I.. 2003. Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J Agric Food Chem. 51:6452–6455. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H.. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Kim SK.. 2004. Physicochemical properties of buckwheat starches from different areas. Kor J Food Sci Technol. 36:598–603. [Google Scholar]
- Kump B, Javornik B.. 1996. Evaluation of genetic variability among common buckwheat (Fagopyrum esculentum Moench) populations by RAPD markers. Plant Sci. 114:149–158. [Google Scholar]
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA.. 2008. . Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale – a wild ancestor of cultivated buckwheat. BMC Plant Biol. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako T, Yamane K, Ohnishi O.. 2002. Two new Fagopyrum (Polygonaceae) species, F. gracilipedoides and F. jinshaense from Yunnan, China . Genes Genet Syst. 77:399–408. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang J, Lu C, Shen Q, Yan Y, Xu C, Song C.. 2015. The complete chloroplast genome sequence of Fagopyrum cymosum. Mitochondrial DNA. 27:2410–2411. [DOI] [PubMed] [Google Scholar]