Abstract
This study reported the complete nucleotide sequence of the Nicotiana tabacum TN90 chloroplast (cp) genome. The cpDNA was 155 992 bp in length and contained 133 individual genes (79 protein encoding genes, 30 tRNA genes and four rRNA genes). Maximum-likelihood (ML) phylogenetic tree for 17 species with Arabidopsis thaliana, Oryza sativa, and Anomochloa marantoidea as an outgroup resulted in a single tree with − lnL =542 222.71, where the Nicotiana tabacum TN90 plastid was clustered with three previous reported Nicotiana species: N. tomentosiformis, N. undulata and N. tabacum. The TN90 variety of tobacco cp genome sequence reported in this study will accelerate tobacco improvement in the future.
Keywords: Chloroplast genome, Nicotiana tabacum TN90, phylogenetic analysis
Tobacco (Nicotiana tabacum) is a model plant organism for studying fundamental biological processes and is one of the most widely cultivated non-food crops worldwide (Peedin 2011). Over 75 naturally occurring Nicotiana species, including 49 native to America and 25 native to Australia (Chase et al. 2003). Most commercial tobaccos cultivated today belong to the species Nicotiana tabacum L., among that 41 600 N. tabacum cultivated varieties (cultivars) are listed in the National Plant Germplasm System (National Plant Germplasm System: http://www.arsgrin.gov/npgs/). Besides, tobacco is also used as a model for plant disease susceptibility, which shares with other Solanaceae plants including potato, tomato and pepper. Diseases affecting tobacco include the tobacco mosaic virus (TMV), the tobacco vein-mottling virus (TVMV), the tobacco etch virus (TEV) and the potato virus Y (PVY).
The TN90 variety of tobacco is a worldwide commercial Burley tobacco variety that has been bred to resist all kinds of viral infections. Besides, the TN90 was also the first variety of tobacco species with a reported nuclear genome sequence (Sierro et al. 2014). In this study, total DNA of the TN90 was sequenced with Illumina HiSeq 2000 (Illumina Inc., San Diego, CA) and about 22 342 988 paired-end sequence reads were produced. Plant sample for this species was collected and conserved in Yunnan Reascend Tobacco Technology (Group) Co., Ltd., Kunming, China. Then the sequence reads were first filtered to previously reported tobacco chloroplast genome: N. tomentosiformis (NC_007602), N. undulata (JN563929) and N. tabacum (NC_001879.2), and assembled with SOAP2 (Li et al. 2009). The finally assembled cpDNA of N. tabacum TN90 plastid is 155 992 bp in length (GenBank accession no. KU199713). Genes encoded by this genome were annotated by using DOGMA (Wyman et al. 2004). The two inverted repeated regions (IRs) of this genome are 25 768 bp, and the large single-copy (LSC) region and small single-copy (SSC) region are 85 987 bp and 18 469 bp, respectively. The overall AT contents were 63.6% and in the LSC, SSC and IR regions, they were 65.9%, 70.8% and 57.8%, respectively. All the 133 individual genes (79 protein encoding genes, 30 tRNA genes and four rRNA genes), 18 genes are duplicated in the IR regions. Among them, 15 genes contained one intron, while three genes had two introns.
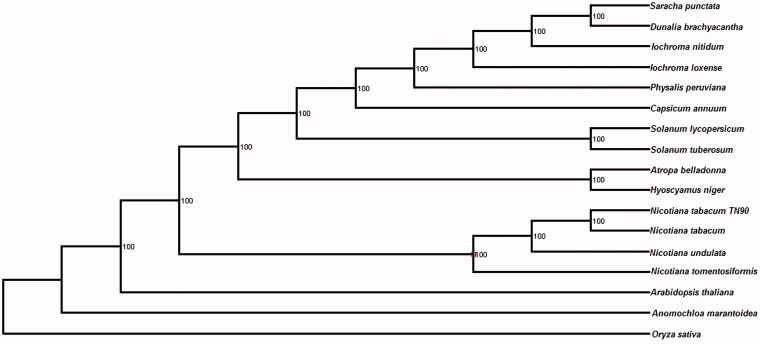
To study its phylogenetic relationships with previously reported cp genome sequences, all 13 complete cp genome sequences of Solanaceae species and three species from other Family were download for analyses. Maximum-likelihood (ML) phylogenetic tree for 17 species with Arabidopsis thaliana, Oryza sativa, and Anomochloa marantoidea as an outgroup resulted in a single tree with − lnL =542 222.71. Bootstrap analyses indicated that all the nodes were supported by values of 100% (Figure 1), where the TN90 plastid was clustered with three previous reported Nicotiana species: N. tomentosiformis, N. undulata and N. tabacum. In all, the TN90 variety of tobacco cp genome sequence reported in this study will accelerate tobacco improvement in the future.
Figure 1.
Phylogenetic relationship of N. tabacum TN90 plastid in the family Solanaceae with previously reported complete cp genomes. The accessions for these species were Anomochloa marantoidea (NC_014062); Arabidopsis thaliana (NC_000932); Atropa belladonna (NC_004561); Capsicum annuum (KJ619462); Dunalia brachyacantha (NC_026906); Hyoscyamus niger (KF248009); Iochroma loxense (NC_026726); Iochroma nitidum (NC_026567); Nicotiana tomentosiformis (NC_007602); Nicotiana undulata (JN563929); Nicotiana tabacum (NC_001879); Oryza sativa Japonica (NC_001320); Physalis peruviana (NC_026570); Saracha punctata (KP280050); Solanum lycopersicum (NC_007898); Solanum tuberosum (DQ386163).
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS.. 2003. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot. (Lond.) 92:107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RQ, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J.. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967. [DOI] [PubMed] [Google Scholar]
- Peedin GF. 2011. Tobacco cultivation In: Myers ML, editor. Specialty crops. Geneva: International Labor Organization. [Google Scholar]
- Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV.. 2014. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 5:3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]