Abstract
The complete mitochondrial genome of the treehopper species, Entylia carinata, was sequenced using the next-generation sequencing method. The completely assembled mitochondrial genome is 15,483 bp long. The genome follows the typical invertebrate mitochondrial gene arrangement, and includes 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one A + T-rich region. Two tandem repeats were also identified within the A + T-rich region.
Keywords: Entylia carinata; Membracidae, mitochondrial genome, tandem repeat
The treehopper family, Membracidae, comprises 400 genera and ∼3500 described species distributed globally (Cryan et al. 2004; Deitz & Wallace 2010, 2012). Membracidae exhibits several remarkable life-history traits, including brood care and ant-tending mutualisms (Deitz & Wallace 2010). However, species-level phylogenies and resources for molecular systematics remain limited for this group. The lack of molecular markers hinders research questions that address species phylogeny, the evolution of life-history traits and ecological adaptations, and historical biogeography. To provide additional membracid resources, we sequenced the complete mt genome of Entylia carinata. This species has a widespread distribution across the North American continent (McKamey 1998; Godoy et al. 2006; Deitz & Wallace 2010). Entylia carinata is a polyphagous phloem-sap feeder that relies on microbial symbionts for nutritional supplementation of its herbaceous plant diet (Buchner 1965; Wood 1993).
Insect specimens were collected in 2013 from Yale Campus, Orange, Connecticut, U.S.A (41°15′16.3″N, 72°59′33.4″W) and deposited in the University of Hawaii Insect Museum (Accession Number: UHIM2016-4430, UHIM2016-4431). Purified genomic DNA was extracted with a Qiagen DNeasy kit and sequenced at the University of Texas, Austin GSAF. Genomic libraries were prepared from 300 base pair (bp) fragments and sequenced on an Illumina MiSeq (2 × 300 bp PE reads). A single full-length contig was obtained for the mt genome with SPAdes V3.6.2 (Bankevich et al. 2012). Completeness was assessed by read mapping with Bowtie2 (Langmead & Salzberg 2012) to confirm that no coverage breaks exist. Protein-coding genes (PCGs), rRNAs, and tRNAs were annotated in Geneious v9.0.2 (Kearse et al. 2012). Gene predictions were verified with related mt genomes (Figure 1). The complete mt genome of E. carinata is 15,483 bp (GenBank no. KX495488) with 168× the average read coverage. We identified 37 genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and the A + T-rich region.
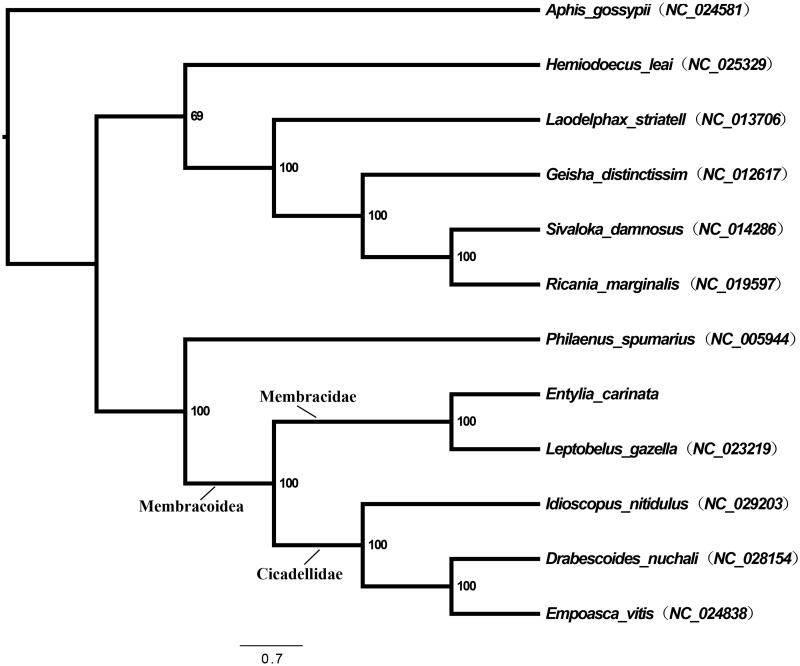
Figure 1.
Maximum-likelihood phylogeny of Hemiptera species with fully sequenced mitochondrial genomes. Phylogenetic reconstructions were done from a concatenated matrix of all 13 protein-coding mitochondrial genes with RAxML-HPC2 under the GTRCAT model in the CIPRES portal (Miller et al. 2010; Stamatakis 2014).
The nucleotide composition of E. carinata has a biased A + T content of 78.1% for the majority strand (J-strand), which is much lower than the related Leptobelus gazella (79.4%; Membracidae) (Zhao & Liang 2016). All 13 PCGs start with ATN codons. Nine PCGs (ATP6, ATP8, CYTB, ND1, ND2, ND3, ND4, ND4L, and ND6) use the typical stop-codons (TAA or TAG), while the remaining four PCGs (COX1, COX2, COX3, and ND5) use the incomplete T stop-codon. The putative A + T-rich region is 1474 bp long (79.4% A + T content). Finally, two tandem repeats were observed in the A + T-rich region, which were shared with L. gazella.
To verify the phylogenetic placement of E. carinata within the Auchenorrhyncha (Hemiptera), a maximum-likelihood phylogeny with 13 PCGs was reconstructed in RAxML-HPC2 (GTRCAT model) in CIPRES (Miller et al. 2010; Stamatakis 2014). Phylogenetic relationships validate the position of E. carinata within the Auchenorrhyncha and also within Membracidae (Figure 1). Furthermore, both E. carinata and L. gazella are placed in a monophyletic group with leafhoppers (Cicadellidae). This result supports previous work indicating the close – possibly paraphyletic – relationship between treehoppers and leafhoppers within Membracoidea (Cryan et al. 2004; Cryan & Urban 2011).
Acknowledgments
We thank Dr. Nancy Moran (University of Texas at Austin) and Ryuchi Koga (AIST, Japan) for their help in finding Entylia carianata populations.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of animals with plant microorganisms: rev. English version. [Translation by Bertha Nueller, with the collaboration of Francis H. Foeckler]. New York: Interscience Publishers. [Google Scholar]
- Cryan JR, Urban JM.. 2011. Higher-level phylogeny of the insect order Hemiptera: is Auchenorrhyncha really paraphyletic. Syst Entomol. 37:7–21. [Google Scholar]
- Cryan JR, Wiegmann BM, Deitz L, Dietrich CH, Whiting MF.. 2004. Treehopper trees: phylogeny of Membracidae (Hemipetera: Cicadomorpha: Membracoidea) based on molecules and morphology. Syst Entomol. 29:441–445. [Google Scholar]
- Deitz LL, Wallace MS.. 2010. Treehoppers: Aetalionidae, Melizoderidae, and Membracidae (Hemiptera). The National Science Foundation, NCSU Insect Museum, and East Stroudsburg University of Pennsylvania, USA. Available from: http://treehoppers.insectmuseum.org.
- Deitz LL, Wallace MS.. 2012. Richness of the Nearctic treehopper fauna (Hemiptera: Aetalionidae and Membracidae). Zootaxa. 3423:1–26. [Google Scholar]
- Godoy C, Miranda X, Nishida K.. 2006. Membrácidos de la América tropical: treehoppers of tropical America. 1st ed. Santo Domingo De Heredia, Costa Rica: Instituto Nacional De Biodiversidad; p. 352. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKamey SH. 1998. Taxonomic catalogue of the Membracoidea (exclusive of leafhoppers): second supplement to fascicle 1 Membracidae of the general catalogue of the Hemiptera. Mem Am Entomol Inst. 6:1–377. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees In: Gateway computing environments workshop (GCE); New Orleans (LA). p. 1–8. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK. 1993. Diversity in the new world Membracidae. Annu Rev Entomol. 38:409–435. [Google Scholar]
- Zhao X, Liang AP.. 2016. Complete DNA sequence of the mitochondrial genome of the treehopper Leptobelus gazella (Membracoidea: Hemiptera). Mitochondrial DNA Part A. 27:3318–3319. [DOI] [PubMed] [Google Scholar]