Abstract
We sequenced almost complete mitochondrial genome of Apis florea (Insecta: Hymenoptera: Apocrita: Apidae) with length of 15,933 bp. The genome has similar codon usage and gene organization to those of mitogenome reported for other Hymenoptera. It includes 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes and a noncoding region with very high AT-base content. Phylogenetic analyses showed that, Apis florea and (Apis cerana+ Apis mellifera) are the sister taxa, and more original, which is widely accepted view.
Keywords: Apis florea, mitochondrial genome, phylogenetic analysis
In this study, we presented the mitochondrial genome of the Apis species, Apis florea, which belong to corbiculate tribes. The species is extending some 7000 km from its eastern-most extreme in Vietnam and south-eastern China, across mainland Asia, along and below the southern flanks of the Himalayas, westwards to the Plateau of Iran and southwesterly into Oman (Hepburn et al. 2005). Specimen in this study was collected on 22 May 2010 in Meng Yang town, Jing Hong City, Yunnan province, southwest China (coordinate as follows: N22°04.070′E100°56.303′), and the specimen is stored in Kunming, Yunnan, China. The almost entire mitochondrial genome of Apis florea was 15,993 bp and deposited in the GenBank under the accession number KC170303 in NCBI. The genome has similar codon usage and gene organization to those of mitogenome reported for other Hymenoptera. It includes 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes and a noncoding region with very high AT-base content. Genetic DNA from honeybee head was individually extracted. Annotation of the complete assembled mitochondrial genome was performed with Dual OrganellarGenoMe Annotator (DOGMA) (Wyman et al. 2004). The total base composition was A (43.46%), T (41.87%), G (5.48%), C (9.19%), suggesting that the percentage of A + T (85.33%) was higher than G + C (14.67%).
The phylogenetic analyses in this study concur with the proposed and widely accepted view of the internal relationships of Apis, A. cerana and A. mellifera are sister taxa and A. florea are more original (Arias and Sheppard 1996, 2005; Ratnieks 2006; Raffiudin and Crozier 2007). The consensus tree topology showed corbiculate bees to be a completely monophyletic taxa (Figure 1) and the relation between tribes was observed having 1000 bootstrap value. The complete mitochondrial genomes of seven other species are available from GenBank and the science name and accession numbers are as follows: Apis cerana (GQ162109), Apis mellifera ligustica (L06178), Melipona bicolour (NC_004529), Bombus hypocrita sapporensis (NC_011923), Bombus ignites (DQ870926), Evania appendigaster (NC_013238).
Figure 1.
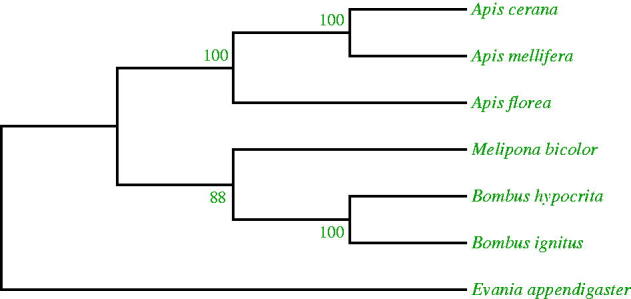
Inferred phylogenetic relationship among corbiculate tribes.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Arias MC, Sheppard WS.. 1996. Molecular phylogenetics of honey bee species (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol Phylogenet Evol. 5:557–566. [DOI] [PubMed] [Google Scholar]
- Arias MC, Sheppard WS.. 2005. Phylogenetic relationships of honey bees (Hymenoptera: Apinae: Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 37:25–35. [DOI] [PubMed] [Google Scholar]
- Hepburn HR, Radloff SE, Otis GW, Fuchs S, Verma LR, Tan K, Chaiyawong T, Tahmasebi G, Wongsiri S.. 2005. Apis florea: morphometrics, classification and biogeography. Apidologie. 36:359–376. [Google Scholar]
- Raffiudin R, Crozier RH.. 2007. Phylogenetic analysis of honey bee behavioral evolution. Mol Phylogenet Evol. 43:543–552. [DOI] [PubMed] [Google Scholar]
- Ratnieks WFL. 2006. Asian honey bees: biology, conservation and human interactions. Nature. 442:249–249. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]


