Abstract
The complete mitochondrial genome of the nine-spined stickleback Pungitius pungitius was obtained with Illumina sequencing of genomic DNA. The assembled mitogenome sequence was 16 582 bp long, and the gene number, order and contents were identical to those of other sequenced Pungitius mitogenomes. The complete mitogenome of P. pungitius from its European range can provide an important template for further phylogenetic and population genetic studies of the Pungitius species complex.
Keywords: Gasterosteidae, genome, mtDNA, nine-spined stickleback, Pungitius pungitius
The nine-spined stickleback Pungitius pungitius is the most geographically widespread member of the genus Pungitius, with a circumpolar distribution (Wootton 1976). It is an important model species in evolutionary biology, genetics and behavioural research (Östlund-Nilsson et al. 2007; Merilä 2013). Pungitius pungitius has been the focus of many population genetic and phylogeographic studies using mitochondrial gene fragments (e.g., Aldenhoven et al. 2010; Shikano et al. 2010; Bae & Suk 2015; Wang et al. 2015), but until now, only a partial assembly of the whole mitochondrial genome has been available from one individual originating from Hokkaido, Japan (Kawahara et al. 2009).
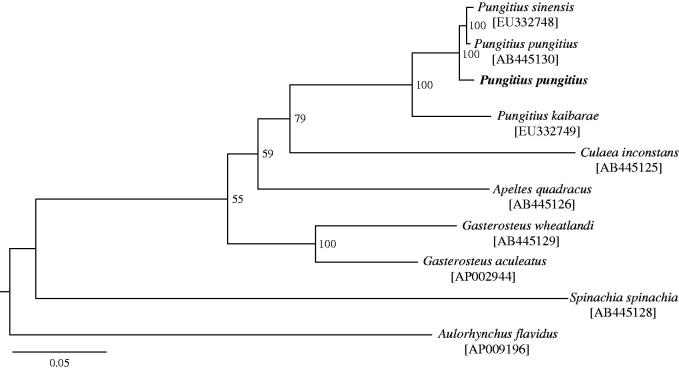
We sequenced the genomic DNA of one P. pungitius individual collected from Montagny-lès-Seurre, France (47°01′ N, 05°15′E) on the Illumina HiSeq2000 platform with 100 paired-end strategy, and aligned 7.5 million reads against the Pungitius sinensis mitogenome (Hwang et al. 2012a) with bwa-0.5.10 (Li & Durbin 2009). The mean coverage of the alignment was 56.89-fold, and 100% of the genome has 1-fold coverage (99.35% with ≥20-fold coverage). The consensus sequence of the P. pungitius mitochondrial genome was exported with SAMtools 1.2 (Li et al. 2009) and manually checked. The complete mitochondrial genome of P. pungitius was 16 582 bp (GenBank Accession No. KT989571) and consisted of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a control region. The order and direction of these genes are identical to those of other Gasterosteidae species including Pungitius fishes (Miya et al. 2001; Kawahara et al. 2009; Hwang et al. 2012a,2012b). Of the 13 protein-coding genes, 4 (ND2, COII, ND4 and Cytb) showed an incomplete stop codon. The base composition of the entire genome was 27.5% for A, 28.0% for T, 17.3% for G and 28.2% for C. The phylogenetic position of P. pungitius among Gasterosteidae fishes was confirmed based on a maximum-likelihood tree constructed with the 37 genes (15 583 bp in total) using RAxML v.8.0 (Stamatakis 2014; GTR + GAMMA model, 37 gene partitions and 100 thorough bootstrap replicates). P. pungitius clustered with other Pungitius fishes, including Japanese P. pungitius, Korean P. sinensis and Korean P. kaibarae (Figure 1). Nucleotide identity in the 37 gene regions of P. pungitius was 98.7%, 98.6% and 93.9% in comparisons with those of Japanese P. pungitius (AB445130), Korean P. sinensis (EU332748) and Korean P. kaibarae (EU332749), respectively.
Figure 1.
A maximum-likelihood tree inferred from 37 mitochondrial genes among nine Gasterosteidae fishes and an out-group species. Bootstrap support is indicated at nodes. GenBank accession numbers are indicated in brackets.
Acknowledgements
We thank Pekka Ellonen, Laura Häkkinen, Tiina Hannunen and Sami Karja for help with laboratory and bioinformatics work. Takahito Shikano is thanked for his help with preparation of this manuscript and Jacquelin DeFaveri for useful suggestion on earlier version of it. The sequencing was conducted at Finnish Institute for Molecular Medicine.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by grants (108601 & 118673) from the Academy of Finland.
References
- Aldenhoven JT, Miller MA, Showers Corneli P, Shapiro MD.. 2010. Phylogeography of ninespine sticklebacks (Pungitius pungitius) in North America: glacial refugia and the origins of adaptive traits. Mol Ecol. 19:4061–4076. [DOI] [PubMed] [Google Scholar]
- Bae HG, Suk HY.. 2015. Population genetic structure and colonization history of short ninespine sticklebacks (Pungitius kaibarae). Ecol Evol. 5:3075–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D-S, Song HB, Lee J-S.. 2012a. Complete mitochondrial genome of the Amur stickleback Pungitius sinensis (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:293–294. [DOI] [PubMed] [Google Scholar]
- Hwang D-S, Song HB, Lee J-S.. 2012b. Complete mitochondrial genome of the Amur stickleback Pungitius kaibarae (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA 23:313–314. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M.. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50:401–404. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilä J. 2013. Nine-spined stickleback (Pungitius pungitius): an emerging model for evolutionary biology research. Ann NY Acad Sci. 1289:18–35. [DOI] [PubMed] [Google Scholar]
- Miya M, Kawaguchi A, Nishida M.. 2001. Mitogenetic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Phylogenet Evol. 18:1993–2009. [DOI] [PubMed] [Google Scholar]
- Östlund-Nilsson S, Mayer J, Huntigford F.. 2007. Biology of the three-spined stickleback. Boca Raton: CRC Press. [Google Scholar]
- Shikano T, Shimada Y, Herczeg G, Merilä J.. 2010. History vs. habitat type: explaining the genetic structure of European nine-spined stickleback (Pungitius pungitius) populations. Mol Ecol. 19:1147–1156. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shikano T, Persat H, Merilä J.. 2015. Mitochondrial phylogeography and cryptic divergence in the stickleback genus Pungitius. J Biogeogr. 42:2334–2348. [Google Scholar]
- Wootton RJ. 1976. The biology of the sticklebacks. New York (NY): Academic Press. [Google Scholar]