Abstract
In this study, a complete chloroplast genome sequence of Artemisia fukudo (Asteraceae family) was characterized by de novo assembly using whole genome sequence data. The chloroplast genome was 151,011 bp in length, comprising a large single-copy region of 82,751 bp, a small single copy region of 18,348 bp and a pair of inverted repeats of 24,956 bp. The genome contained 80 protein-coding genes, 4 rRNA genes and 30 tRNA genes. Phylogenetic tree revealed that A. fukudo was closely located in other Artemisia species, Artemisia montana and Artemisia frigida.
Keywords: Artemisia fukudo, chloroplast, genome sequence
The Artemisia genus belongs to the Asteraceae family and is a native to Europe, North Africa, and Asia, including Korea (Abad et al. 2012). In Korea, about 20 Artemisia species were found and have been used as ingredients for food and as medicinal herbs for a long time (Kim et al. 2007). A. fukudo is mainly distributed along the shorelines of Jeju Island, South Korea, and have been used as a flavouring agent and cosmetic ingredient in Korea (Abad et al. 2012). Previous study reported that A. fukudo possessed essential oils composed of oxygenated monoterpene and sesquiterpene hydrocarbons with anti-inflammatory effect, and confirmed its potential herbal medicinal activity (Yoon et al. 2010). Lots of Artemisia species grow together in wild and there is some difficulty to authenticate each species because of their similar morphological traits. Practical barcode markers are necessary to authenticate related species from the processed products (Jung et al. 2014) because dried plant leaves of Artemisia species are used as crude drug. Although complete chloroplast genome sequences of two Artemisia species (JX293720 and KF887960) are known, there was no report for A. fukudo. We developed chloroplast genome sequence of A. fukudo to further apply developing authentication as well as in improving its quality as medicinal plant.
Chloroplast genome sequence of A. fukudo was generated using de novo assembly method for chloroplast genome reported by Kim et al (2015a and 2015b). Artemisia fukudo was collected from Jeungdo, Jeollanam-do, South Korea, and maintained in Hantaek Botanical Garden (http://www.hantaek.co.kr/). We isolated DNA from leaf tissues and performed whole genome shotgun sequencing using an Illumina Miseq platform (Illumina, San Diego, CA). High-quality paired end reads of 1.2 Gb were assembled using CLC genome assembler 4.6 (CLC Inc., Aarhus, Denmark) and contigs representing chloroplast sequence were retrieved, ordered, and combined into a single sequence, by the guidance of chloroplast genome of A. frigida (JX293720) (Liu et al. 2013).
Complete chloroplast genome of A. fukudo (Accession number: KU360270) was 151,011 bp in length, consisted of four distinct parts including a large single copy region of 82,751 bp, a small single copy region of 18,348 bp, and a pair of inverted repeats of 24,956 bp. In genome, a total 114 coding regions, which were comprised 80 protein-coding regions, 4 rRNA genes, and 30 tRNA genes, were predicted through annotation using DOGMA (http://dogma.ccbb.utexas.edu) and BLAST searches.
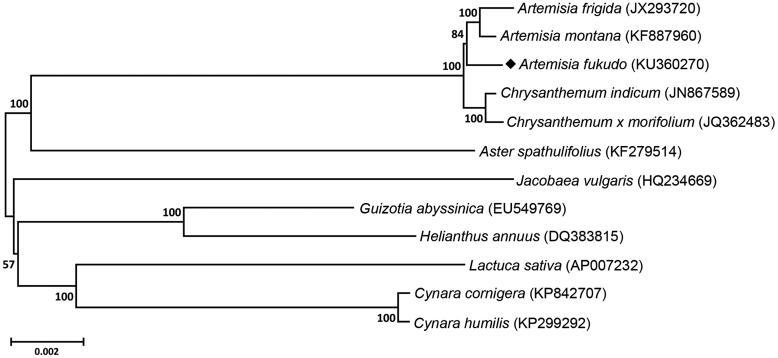
Phylogenetic relationship was analyzed with the chloroplast genome of A. fukudo along with 11 species belonging to the Asteraceae family. A total of 74 coding sequences were used for the analysis, and phylogenetic tree was constructed using neighbour-joining method in the MEGA 6.0 (Tamura et al. 2013). Phylogenetic tree revealed that A. fukudo was grouped with two Artemisia (A. montana and A. frigida), as expected. However, Guizotia abyssinica and Helianthus annuus, which belong to the subfamily Asteroideae, were placed with Lactuca sativa (Subfamily Cichorioideae) and two Cynara species (Subfamily Carduoideae), suggesting that there is still a contradiction in taxonomical classification of Asteraceae family (Xia et al. 2015) (Figure 1).
Figure 1.
Phylogenetic tree revealing the relationship of A. fukudo with 11 species belonging to the Asteraceae family. This tree were constructed with those complete chloroplast genome sequences using neighbour-joining method with 1000 bootstrap values in the MEGA 6, and numbers above each node indicate the bootstrap values.
Acknowledgments
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by ‘‘15172MFDS246’’ from Ministry of Food and Drug Safety in 2015, Republic of Korea.
References
- Abad MJ, Bedoyq LM, Apaza L, Bermejo P.. 2012. The Artemisia L. genus: a review of bioactive essential oils. Molecules. 17:2542–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim K, Yang K, Bang KH, Yang TJ.. 2014. Practical application of DNA markers for high throughput authentication of Panax ginseng and Panax quinquefolius from commercial ginseng products. J Ginseng Res. 38:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Lee JA, Yoon WJ, Kim JY, Song GP, Park SY.. 2007. The cytotoxicity of Artemisia fukudo extracts against HL-60 cells. J Korean Soc Food Sci Nutr. 36:819–824. [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ.. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. . 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ.. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 8:e57533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 . Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon WJ, Moon J, Song G, Lee Y, Han M, Lee J, Ihm B, Lee W, Lee N, Hyun C.. 2010. . Artemisia fukudo essential oil attenuates LPS-induced inflammation by suppressing NF-kappaB and MAPK activation in RAW 264.7 macrophages. Food Chem Toxicol. 48:1222–1229. [DOI] [PubMed] [Google Scholar]
- Xia Y, Hu Z, Li X, Wang P, Zhang X, Li Q, Lu C.. 2015. The complete chloroplast genome sequence of Chrysanthemum indicum. Mitochondrial DNA. DOI: 10.3109/19401736.2015.1106494. [DOI] [PubMed] [Google Scholar]