Abstract
In this study, we report the complete mitochondrial genome of Hirsutella rhossiliensis (Ophiocordycipitaceae, Hypocreales, Ascomycota). We construct the mitochondrial DNA genome organization of 62 483 bp in length of H. rhossiliensis by using the whole-genome resequencing method. Conserved genes including the large and small rRNA subunits, 26 tRNA and 14 protein-coding genes are identified. These protein-coding genes utilize ATG, GTG or TTG as initiation codons and TAA or TAG as termination codons. Moreover, we detect 10 group I introns and one unclassified intron in six genes (rnl, cob, cox1, cox3, nad1 and nad5) encoding ORFs of ribosomal protein S3 and GIY-YIG/LAGLIDADG endonucleases or hypothetical proteins. This mitochondrial genome will be useful in understanding the distribution and genetic diversity of this species.
Keywords: Hirsutella rhossiliensis, Hypocreales, mitochondrial genome, Ophiocordycipitaceae
Hirsutella rhossiliensis and Hirsutella minnesotensis (Ophiocordy-cipitaceae, Hypocreales, Ascomycota) are two representatives of nematode endoparasitic fungi (Liu et al. 2009; Sun et al. 2015). The complete mitogenome of H. minnesotensis has been reported (Zhang et al. 2015). Here, we present the complete mitogenome of H. rhossiliensis strain USA-87-5 (GenBank accession no. KU203675) isolated from parasitized second-stage juveniles of Heterodera glycines from a soybean field in Cottonwood county, Minnesota, America (44°2′24″ N, 94°55′48″ W). The specimen was deposited in the Herbarium of Microbiology, Academia Sinica (HMAS), while living culture was deposited at the China General Microbiological Culture Collection Center (CGMCC) (HMAS 246731; CGMCC 3.17882).
Whole-genome resequencing is performed on an Illumina HiSeq 2500-PE125 platform (Illumina Inc., San Diego, IL). A lane of 2 × 125 bp paired-end resequencing creates 3 915 863 000 clean reads based on our DNA sample. These reads are mapped with the Burrows-Wheeler Aligner (version 0.7.0) (Li & Durbin 2009) to the reference genome of H. rhossiliensis (Lai et al. unpublished) and assembled using SPAdes 3.1.1 (Bankevich et al. 2012) into 162 716 contigs. Then BLAST searches against known complete mitogenome of H. minnesotensis suggest 10 high similar fragments of total 49 257 bp with circularity. These fragments are annotated as two rRNA, 26 tRNA and 14 standard protein-coding genes using the MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/MfannotInterface.pl). Gaps are filled by general PCR using a pair of specific primers designed by the software Primer3 (http://frodo.wi.mit.edu/primer3/) according to known flanking sequences.
The length of complete mitochondrial genome of H. rhossiliensis is determined to be 62 483 bp and contains 26 tRNAs, two rRNAs and 14 protein-coding sequences. The nucleotide composition of H. rhossiliensis is 36.5% A, 12.8% C, 15.4% G and 35.3% T. The arrangement of 14 protein-coding genes and rRNAs is followed as rnl, nad2, nad3, atp9, cox2, nad4L, nad5, cob, cox1, nad1, nad4, atp8, atp6, rns, cox3 and nad6 identical to that of other common fungal mitochondrial genomes. Structural genes including 14 protein-coding genes, two rRNA and 26 tRNA genes cover 61.1% (38 180 bp) of the mitochondrial genome. The intergenic sequences have a total length of 24 303 bp occupying 38.9% of the genome. Intron sequences including introns of protein-coding genes account for 19.8% (12 395 bp) of the mitogenome.
The set of 26 tRNA genes codes for all 20 standard amino acids. Seventeen tRNA genes are adjacent to rnl, four tRNA genes approach to rns and five tRNA genes locate around three protein-coding genes (cob, cox1 and cox3). Twelve introns invade six genes including rnl (one), cob (three), cox1 (three), cox3 (two), nad1 (one) and nad5 (two). These introns mainly belong to group I introns, but one (i.e. nad5-i1) is unclassified. Intronic proteins include ribosomal protein S3 and GIY-YIG/LAGLIDADG-type endonucleases or hypothetical proteins.
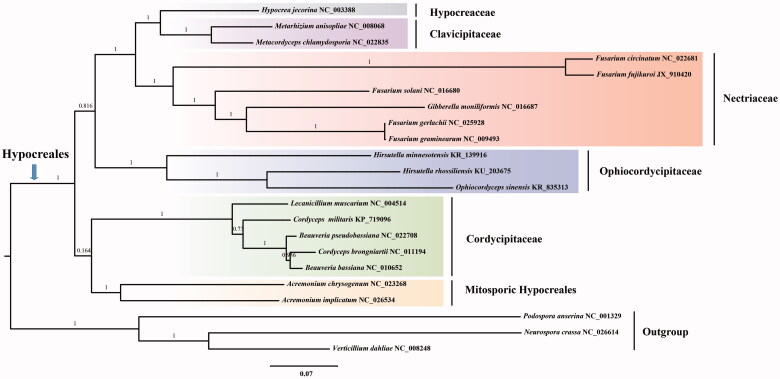
Phylogenetic analysis based on whole mitogenome sequences confirms H. rhossiliensis as a member of the fungal order Hypocreales. Hirsutella rhossiliensis is clustered together with H. minnesotensis and Ophiocordyceps sinensis within the family Ophiocordycipitaceae according to our phylogenetic analysis (Figure 1), with consistent taxonomic status according to phylogenetic analysis of nuclear genes of Hypocreales (Sung et al. 2007).
Figure 1.
Phylogenetic analysis based on neighbour-joining method implemented in FastTree (Price et al. 2009) among 19 taxa of Hypocreales using whole mitogenome sequences. They are currently available in the GenBank database. The support values were shown above the nodes. Podospora anserine, Neurospora crassa, and Verticillium dahliae were used as the outgroups. Note that the accession number of Ophiocordyceps sinensis has not yet been released. The complete mitochondrial genome of O. sinensis has been reported (Li et al. 2015).
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing. This work was supported by the National Key Basic Research Program of China (Program no. 973, Grant no. 2013CB127506), the National Natural Science Foundation of China (Grant no. 30800732).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu XD, Yang RH, Hsiang T, Wang K, Liang DQ, Liang F, Cao DM, Zhou F, Wen G, et al. 2015. Complete mitochondrial genome of the medicinal fungus Ophiocordyceps sinensis. Sci Rep. 5:13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Xiang MC, Che YS.. 2009. The living strategy of nematophagous fungi. Mycoscience 50:20–25. [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JZ, Park SY, Kang S, Liu XZ, Qiu JZ, Xiang MC.. 2015. Development of a transformation system for Hirsutella spp. and visualization of the mode of nematode infection by GFP-labeled H. minnesotensis. Sci Rep. 5:10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW.. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 57:5–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Zhang S, Liu XZ.. 2015. The complete mitochondrial genome of the nematode endoparasitic fungus Hirsutella minnesotensis. Mitochondrial DNA. doi: 10.3109/19401736.2015.1046126. [DOI] [PubMed] [Google Scholar]