Abstract
Gyrodactylus gurleyi, was inhabited on the fins and gills of goldfish (Carassius auratus), which belonged to the family Gyrodactylidae. In this study, we sequenced the complete mitochondrial genome of G. gurleyi with the total length of 14 771 bp. The mitogenome contained 12 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and two major non-coding regions (NC1 and NC2). The overall AT content was 72.1%. In phylogenetic analysis, G. gurleyi and G. kobayashii clustered together and then united with the clade of other three Gyrodactylus species (G. salaris, G. thymalli and G. derjavinoides) with high nodal support.
Keywords: Gyrodactylus gurleyi, Gyrodagtylidae, mitochondrial genome, phylogenetics
Gyrodactylus gurleyi was collected on the fins and gills of goldfish (Carassius auratus) from Wuhan (30°31′23′′N, 114°23′01′′E), China. It was identified by morphology and ITS molecular marker (Li et al. 2013). The specimen (accession no. IHB20150315006) was stored in the Museum of Aquatic Organisms, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China. The sequences of total mt genomic DNA of G. gurleyi were retrieved using long PCR and Sanger method of DNA sequencing.
The complete mt genome of G. gurleyi was circular and 14 771 bp in size (GenBank accession no. KU659806). It contained 12 protein-coding genes (PCGs, lacking Atp8), 22 tRNA genes, two rRNA genes and two major non-coding regions (NC1 and NC2) (Table 1). All the genes were transcribed from the same strand. The nucleotide composition was computed at 29.2%A, 16.9% G, 42.9% T and 11.1% C. The AT content of 72.1% was lower than that of Paragyrodactylus variegatus (76.3%), but higher than that of other four Gyrodactylus species (G. salaris, 62.3%; G. thymalli, 62.8%; G. derjavinoides, 68.2%; G. kobayashii, 71.6%) (Huyse et al. 2007; Plaisance et al. 2007; Huyse et al. 2008; Ye et al. 2014; Zhang et al. 2016).
Table 1.
Organization of the mitochondrial genome of Gyrodactylus gurleyi.
| Position |
Intergenic | Codon |
|||||
|---|---|---|---|---|---|---|---|
| Gene/region | From | To | Size | nucleotides | Start | Stop | Anti-codon |
| Cox3 | 1 | 639 | 639 | ATG | TAA | ||
| tRNA-His | 643 | 707 | 65 | 3 | GTG | ||
| Cytb | 711 | 1784 | 1074 | 3 | ATG | TAA | |
| Nad4l | 1784 | 2032 | 249 | −1 | ATG | TAA | |
| Nad4 | 2005 | 3213 | 1209 | −28 | ATG | TAA | |
| tRNA-Phe | 3216 | 3281 | 66 | 2 | GAA | ||
| NC1 | 3282 | 4064 | 783 | ||||
| Atp6 | 4065 | 4577 | 513 | ATG | TAA | ||
| Nad2 | 4587 | 5444 | 858 | 9 | ATG | TAA | |
| tRNA-Val | 5449 | 5513 | 65 | 4 | TAC | ||
| tRNA-Ala | 5514 | 5581 | 68 | TGC | |||
| tRNA-Asp | 5584 | 5648 | 65 | 2 | GTC | ||
| Nad1 | 5649 | 6536 | 888 | ATG | TAA | ||
| tRNA-Asn | 6538 | 6603 | 66 | 1 | GTT | ||
| tRNA-Pro | 6604 | 6667 | 64 | TGG | |||
| tRNA-Ile | 6663 | 6729 | 67 | −5 | GAT | ||
| tRNA-Lys | 6730 | 6793 | 64 | CTT | |||
| Nad3 | 6797 | 7144 | 348 | 3 | ATG | TAG | |
| tRNA-Ser(AGN)(S1) | 7145 | 7203 | 59 | GCT | |||
| tRNA-Trp | 7207 | 7271 | 65 | 3 | TCA | ||
| Cox1 | 7276 | 8823 | 1548 | 4 | ATG | TAA | |
| tRNA-Thr | 8836 | 8900 | 65 | 12 | TGT | ||
| rrnL | 8900 | 9853 | 954 | −1 | |||
| tRNA-Cys | 9854 | 9914 | 61 | GCA | |||
| rrnS | 9915 | 10,624 | 710 | ||||
| Cox2 | 10,625 | 11,206 | 582 | ATG | TAA | ||
| tRNA-Glu | 11,319 | 11,389 | 71 | 112 | TTC | ||
| Nad6 | 11,393 | 11,875 | 483 | 3 | ATG | TAG | |
| tRNA-Tyr | 11,884 | 11,951 | 68 | 8 | GTA | ||
| tRNA-Leu(CUN)(L1) | 11,954 | 12,019 | 66 | 2 | TAG | ||
| tRNA-Gln | 12,027 | 12,089 | 63 | 7 | TTG | ||
| tRNA-Met | 12,091 | 12,157 | 67 | 1 | CAT | ||
| NC2 | 12,158 | 12,940 | 783 | ||||
| tRNA-Ser(UCN)(S2) | 12,941 | 12,998 | 58 | TGA | |||
| tRNA-Leu(UUR)(L2) | 12,999 | 13,066 | 68 | TAA | |||
| tRNA-Arg | 13,071 | 13,141 | 71 | 4 | TCG | ||
| Nad5 | 13,140 | 14,690 | 1551 | −2 | ATG | TAG | |
| tRNA-Gly | 14,702 | 14,768 | 67 | 11 | TCC | ||
| 14,772 | 14,771 | 3 | |||||
The length of 12 PCGs was 9942 bp, accounting for 67.3% of the full length of the genome. As the four Gyrodactylus species (G. salaris, G. thymalli, G. derjavinoides and G. kobayashii), ATG was the unique start codon. The stop codon TAG was only found in three PCGs (Nad3, Nad5 and Nad6), whereas TAA in the rest of the PCGs. The size of the 22 tRNA genes was 1374 bp, varying from 58 bp (tRNASer(AGN)) to 71 bp (tRNAGlu). Nineteen of them had conventional secondary structure, while tRNASer(AGN), tRNASer(UCN) and tRNACyslacked DHU arms, which was similar to the other known Gyrodactylidae species. The rrnL and rrnS were 954 bp and 710 bp in size, respectively. They were adjacent to tRNAThr (upstream) and Cox2 (downstream), and separated by tRNACys, as described with other reported monopisthocotyleans (Huyse et al. 2007; Plaisance et al. 2007; Huyse et al. 2008; Perkins et al. 2010; Ye et al. 2014; Zhang et al. 2014a, 2014b, 2016). In addition, there were 37 bp overlapping sequences and 1763 space sequences, among which the biggest were NC1 (783 bp) and NC2 (783 bp).
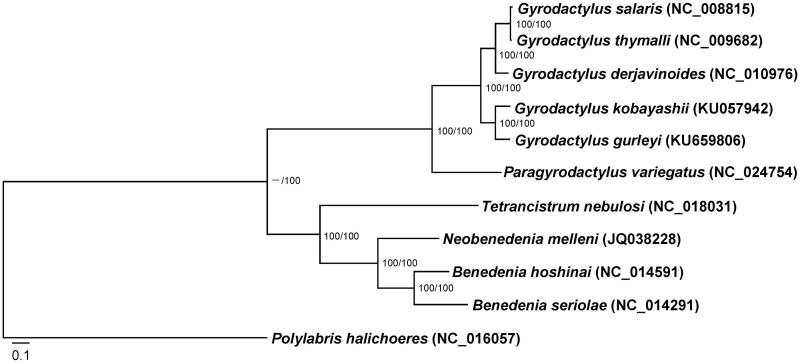
Phylogenetic relationships between G. gurleyi and other nine monopisthocotyleans were inferred by using concatenated amino acid sequences of the 12 PCGs. The same tree topology was obtained by two different computational algorithms: Bayesian inference (BI) and maximum likelihood (ML), in which G. gurleyi and G. kobayashii clustered together and then united with the clade of 3 Gyrodactylus species (G. salaris, G. thymalli and G. derjavinoides) with high nodal support (Figure 1). In addition, although G. gurleyi and G. kobayashii generally parasitized on the same host, sequence alignments showed that 99% (14 623 bp) mitogenome sequence of G. gurleyi was covered by that of G. kobayashii with only 80% identity. In contrast, G. salaris from Atlantic salmon and G. thymalli from grayling share 98% identity with 100% mitogenome alignment coverage.
Figure 1.
Phylogenetic relationships between Gyrodactylus gurleyi and other 9 monopisthocotyleans based on 3044 concatenated amino acid sequences representing 12 mitochondrial protein-coding genes, with Polylabris halichoeres used as an outgroup. The MtZoa model for maximum-likelihood analysis and MtREV model for Bayes analysis are selected. Scale bar corresponds to the estimated number of substitutions per site. Bootstrap support values in percent units above nodes are displayed as follows: maximum likelihood bootstrap/Bayesian posterior probabilities.
Acknowledgments
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the manuscript.
Funding information
This work was supported by the Earmarked Fund for China Agriculture Research System (CARS-46-08), the National Natural Science Foundation of China (31272695, 31572658) and the major scientific and technological innovation project of Hubei Province (2015ABA045).
References
- Huyse T, Buchmann K, Littlewood DTJ.. 2008. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: Monogenea) – a mitogenomic approach for Gyrodactylus species and strain identification. Gene. 417:27–34. [DOI] [PubMed] [Google Scholar]
- Huyse T, Plaisance L, Webster BL, Mo TA, Bakke TA, Bachmann L, Littlewood DTJ.. 2007. The mitochondrial genome of Gyrodactylus salaris (Platyhelminthes: Monogenea), a pathogen of Atlantic salmon (Salmo salar). Parasitology. 134:739–747. [DOI] [PubMed] [Google Scholar]
- Li RR, Li WX, Wu XD, Wang GT.. 2013. Identification of Gyrodactylus species in goldfish (Carassius auratus) through morphological study and the analysis of the rDNA ITS sequence. Acta Hydrobiol Sin. 38:903–909. [Google Scholar]
- Perkins EM, Donnellan SC, Bertozzi T, Whittington ID.. 2010. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int J Parasitol. 40:1237–1245. [DOI] [PubMed] [Google Scholar]
- Plaisance L, Huyse T, Littlewood DTJ, Bakke TA, Bachmann L.. 2007. The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus). Mol Biochem Parasitol. 154:190–194. [DOI] [PubMed] [Google Scholar]
- Ye F, King SD, Cone DK, You P.. 2014. The mitochondrial genome of Paragyrodactylus variegatus (Platyhelminthes: Monogenea): differences in major non-coding region and gene order compared to Gyrodactylus. Parasit Vectors. 7:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zou H, Zhou S, Wu SG, Li WX, Wang GT.. 2016. The complete mitochondrial genome of Gyrodactylus kobayashii (Platyhelminthes: Monogenea). Mitochondrial DNA Part B: Resoureces. 1:154–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu X, Li Y, Xie M, Li A.. 2014a. The complete mitochondrial genome of Tetrancistrum nebulosi (Monogenea: Ancyrocephalidae). Mitochondrial DNA. 1736:1–2. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wu X, Li Y, Zhao M, Xie M, Li A.. 2014b. The complete mitochondrial genome of Neobenedenia melleni (Platyhelminthes: Monogenea): mitochondrial gene content, arrangement and composition compared with two Benedenia species. Mol Biol Rep. 41:6583–6589. [DOI] [PubMed] [Google Scholar]