Abstract
Glioblastoma (GBM) is the most aggressive brain cancer and its relapse after surgery, chemo and radiotherapy appears to be led by GBM stem cells (GSCs). Also, tumor networking and intercellular communication play a major role in driving GBM therapy-resistance. Tunneling Nanotubes (TNTs), thin membranous open-ended channels connecting distant cells, have been observed in several types of cancer, where they emerge to drive a more malignant phenotype. Here, we investigated whether GBM cells are capable to intercommunicate by TNTs. Two GBM stem-like cells (GSLCs) were obtained from the external and infiltrative zone of one GBM from one patient. We show, for the first time, that both GSLCs, grown in classical 2D culture and in 3D-tumor organoids, formed functional TNTs which allowed mitochondria transfer. In the organoid model, recapitulative of several tumor's features, we observed the formation of a network between cells constituted of both Tumor Microtubes (TMs), previously observed in vivo, and TNTs. In addition, the two GSLCs exhibited different responses to irradiation in terms of TNT induction and mitochondria transfer, although the correlation with the disease progression and therapy-resistance needs to be further addressed. Thus, TNT-based communication is active in different GSLCs derived from the external tumoral areas associated to GBM relapse, and we propose that they participate together with TMs in tumor networking.
Keywords: cancer, cell communication, glioblastoma, stem cells, tunneling nanotubes
Introduction
Glioblastoma (GBM) is the most common and aggressive brain cancer which nowadays lacks understanding and resolutive therapeutic strategies. After surgery, patients undergo a mixture of chemo and radiotherapy [1], aiming to kill the remaining cancer cells at the edges of the resected region. Although these treatments have been proven to be effective in extending patients' survival [2], lethal relapse from these peripheral regions occurs in 100% the cases. The elevated intra-tumoral heterogeneity seems to be at the origin of the relapse and particularly due to the presence of GBM stem cells (GSCs) that have been found to be the most resistant to treatments [3–6]. Moreover, it has been shown that post-surgical treatments can induce cellular plasticity and trans-differentiation resulting in more aggressive phenotypes [7]. How this occurs is still not clear, however it appears that intercellular communication in the tumoral context has a major role in the plasticity, survival and progression of many different types of cancer [8,9]. In particular, in the case of GBM, Winkler and colleagues have shown that patient-derived GSCs, xenografted into murine brains, are able to grow tumors where cells interconnect through membranous extensions and form a unique communicating network [10]. These protrusions called Tumor Microtubes (TMs) range in the microscale for their diameter (>1 µm) and could extend for over 500 µm in length, creating a complex tumor cell network. TMs allow the propagation of ion fluxes through GAP-junctional proteins, such as Connexin43 (Cx43), providing a fast, neurite-like, communication between cancer cells. These extensions could also drive the repopulation of surgically injured areas [11]. TM-connected cells resulted to be resistant to chemo and radiotherapy, and the resistance was lost following the inhibition of TM-inducers such as Cx43, GAP43 and TTYH [10–12].
Another mechanism of intercellular communication that has been recently proposed to facilitate tumor progression is represented by Tunneling Nanotubes (TNTs) [13,14]. TNTs are thin cellular extensions connecting distant cells observed in a wide variety of cellular and murine models as well as in ex vivo resections from human tumoral tissue [13]. They are membranous structures supported by an actin-based cytoskeleton and, differently from other cellular protrusions, including TMs (assumed to provide communication through GAP-junctions), are open at both extremities, thus allowing cytoplasmic continuity between connected cells [15,16]. TNTs allow the transfer of various-sized cargos, such as small molecules (e.g. Ca2+ ions), macromolecules (e.g. proteins, nucleic acids) and even organelles (vesicles, mitochondria, lysosomes, autophagosomes, etc.) [17]. They appear to play a critical role in several physio-pathological contexts, as in the spreading of protein aggregates in various neurodegenerative diseases [18–22] or in the transmission of bacteria [23] and viruses [24,25] and, possibly, during development [26]. Functional TNTs have been shown in a variety of cancers using in vitro and ex vivo models [13] where they could be exploited as route for the exchange of material between cancer cells or with the tumoral microenvironment. As consequence of this transfer, cells can acquire new abilities as enhanced metabolic plasticity, migratory phenotype, angiogenic ability and therapy-resistance. In particular, the transfer of mitochondria has been related to all the previously mentioned features since they can provide energy and metabolic support to the cancer cells in displaying their aggressive features as observed in various cancers [14,27].
Few studies have reported TNT-like communication in GBM cells lines [28–30], suggesting that their presence and functionality could be induced/affected by the treatments, contributing to the tumoral progression and treatment-resistance [31,32]. However, no data on the role of TNTs are available in the context of a whole GBM tumor or in primary GSCs. This is likely due to the fragility of these connections and to the low-resolution images that can be obtained in the in vivo studies [10]. Whether in GBM intercellular communication is orchestrated exclusively by TMs or whether TNTs are also present and functional is still not known. Here, we investigate for the first time if TNTs can be formed between patient-derived GSCs and be exploited for exchange cargos using a quantitative approach. We used GSCs derived from the infiltrative region of the tumor, responsible for GBM relapse, thus representing a relevant model for the progression of the disease. In these cells we addressed TNT presence and functionality in both classical adherent cell culture as well as in 3D-tumor organoids as well as the effect of radiotherapy on the TNT-mediated communication.
Material and methods
Cell culture
GBM samples were processed as described by Avril et al. [33]. GSLCs were cultured in suspension in DMEM-F12 (Sigma) supplemented with B27 (50× Gibco), N2 (100× Gibco) and 20 ng/ml of FGF-2 and EGF (Peprotech) at 37°C in 5% CO2 humidified incubators. Fresh medium was added to the cell culture every 2–3 days. All GSLCs were used for the experiments in this medium at less than 25 passages. Absence of alteration upon culture passages on the stemness phenotype was monitored by RT-qPCR. Absence of mycoplasma contamination was verified with MycoAlertTM Mycoplasma Detection Kit (Lonza). All methods were carried out in accordance with the approved guidelines of our institution.
Lentivirus preparation and transduction
Lentiviral particles were produced in human 293T cultured in Dulbecco's Modified Eagle's Medium (ThermoFisher), supplemented with 10% Fetal Bovine Serum (EuroBio) and 1% Pen/Strep (100× Gibco) at 37°C in 5% CO2 humidified incubators. Cells were plated at a 50–70% confluency the day before the transfection. Plasmids coding for lentiviral components, pCMVR8,74 (Gag-Pol-Hiv1) and pMDG2 (VSV-G) vectors, and plasmid of interest at a ratio of 4 : 1 : 4, respectively were transfected using FuGENE HD Transfection reagent according to manufacturer's protocol. MitoGFP (pLV-CMV-mito-GFP) and mCherry (pLV-CMV-mCherry) plasmids encode respectively for a fragment of the subunit VIII of human cytochrome C oxidase fused with GFP, and for cytosolic mCherry under the Cytomegalovirus (CMV) promoter. Viral particles were concentrated using LentiX-Concentrator (TakaraBio) after 48 h, and GSLCs were infected and tested for the expression of the fluorescent marker by flow cytometry at different time points to monitor expression stability.
Tumor organoids preparation and culture
Tumor organoids were prepared according to the protocol published in Hubert et al. [34] and cultured in Neurobasal medium (ThermoFisher) supplemented with B27 (50× Gibco), N2 (100× Gibco), 1% Pen/Strep (100× Gibco), 2 mM L-Glutamine (100× Gibco), 20 ng/ml of FGF, 20 ng/ml EGF Peprotech) and 1.6 ml of GelTrex (ThermoFischer) at 37°C in 5% CO2 humidified incubators up to 23 days. Part of the cultured medium was removed and replaced with fresh one every 2–3 days.
TNT counting
TNTs were identified according to the protocol of Abounit et al. [35]. Cells were plated at the ideal cell density for the observation of TNTs (40 000 cells/cm2 for both GSLCs). The adhesion surface was previously coated with laminin 10 µg/ml (Sigma) for at least 2 h. GSLCs were fixed after 6 h, to avoid excessive cell flattening on the coated surface. Fifteen minutes fixation in solution 1 (2% PFA, 0.05% glutaraldehyde and 0.2 M HEPES in PBS) followed by other 15 min in solution 2 (4% PFA and 0.2 M HEPES in PBS) were performed at 37°C in order to preserve TNTs integrity [35]. Plasma membrane was labeled with fluorescent Wheat Germ Agglutinin (1 : 500 in PBS, Life Technologie) for 20 min at RT. Nuclei were stained with DAPI (1 : 5000 Sigma–Aldrich) before mounting with Mowiol.
Tile confocal images were acquired with a Zeiss LSM 700 controlled by ZEN software. Optimal image stack was applied. The whole volume of the cells was acquired. All the images were processed using ICY software in order to manually count the number of TNT-connected cells. Cells connected through thin, continuous, phalloidin-positive connections were counted as TNT-connected cells.
Immunofluorescence
Cells were seeded on glass coverslips at the TNT-density previously mentioned. Coverslips were coated with 10 µg/ml laminin (Sigma). Cells were fixed with 4% PFA for 20 min at RT. Quenching and permeabilization steps were performed using 50 mM NH4Cl solution and 0.1–0.2% Triton-X100, respectively. Primary antibodies were incubated in 10% FCS-containing PBS solution for 1 h. Anti-αTubulin (1 : 1000 Sigma–Aldrich T9026), anti-TOM20 (1 : 500 Santa Cruz Biotechnology sc-11415) and anti-GAP43 (1 : 500 Cell signaling 8945S) were used. Cells were incubated for 45 min with secondary antibody anti-mouse and anti-rabbit Invitrogen Alexa 488, 564 or 647 antibodies (1 : 1000) or Rhodamine-conjugated Phalloidin (1 : 200, R415 Invitrogen). DAPI (1 : 5000 Sigma–Aldrich) in PBS solution was applied for 5 min before washes and mounting with Mowiol.
Organoids were fixed with a solution of 4% PFA for 1 h at 37°C, washed with PBS-0.5% Tween and incubated in a solution of PBS + 10% FBS + 0.3% BSA (Sigma–Aldrich) + 0.3% Triton-X100 containing primary antibody (mentioned above) overnight at 4°C. Organoids were incubated in the same solution with the corresponding secondary antibody or Rhodamine-conjugated Phalloidin overnight at 4°C, incubated with DAPI (1 : 1000 Sigma–Aldrich) over 6 h and finally mounted with a solution of 70% Glycerol.
Immunofluorescence stainings were analyzed on a Zeiss LSM 700 inverted confocal microscope (Carl Zeiss, Germany), with a Pln-Apo 10×/0.45 to image the entire organoid, 40×: EC Pln-Neo 40×/1.3 (NA = 1.3, working distance = 0.21 mm) or Pln-Apo 63×/1.4 (NA = 1.4, working distance = 0.19 mm) oil lens objective and a camera (AxioCam MRm; Carl Zeiss).
Time-lapse microscopy
Time-lapse microscopy imaging in 2D- and 3D-conditions was performed on an inverted Spinning Disk microscope (Elipse Ti microscope system, Nikon Instruments, Melville, NY, U.S.A.) using 60× 1.4NA CSU oil immersion objective lens using Bright field and Laser illumination 488. Pairs of images were captured in immediate succession with one of two cooled CCD cameras, which enabled time intervals between 20 and 30 s per z-stack. For live cell imaging, the 37°C temperature was controlled with an Air Stream Stage Incubator, which also controlled humidity. Cells were plated in Ibidi µ-Dish 35 mm and incubated with 5% CO2 during image acquisition. Image processing and movies were realized using MetaMorph, FIJI and Imaris software. Time-lapse movies of mitochondria trafficking were created using ImageJ/Fiji software.
Quantification of mitochondria transfer by flow cytometry
Transfer assays were performed according to the protocol of Abounit et al. [35]. Stable GSLC population expressing respectively MitoGFP were used as donor cells and mCherry as acceptor cells and mixed in a 1 : 1 ratio. For the 2D co-culture, cells were plated at the density previously mentioned (see TNT counting). Cells were detached after 2 or 5 days of co-culture with StemPro Accutase (Thermofisher), experimental duplicates were performed for each timepoint and each condition. To monitor the transfer by secretion in 2D co-culture, donor and acceptor cells were co-cultured separated by a 1 µm filter. For tumor organoids, donor and acceptor cells were mixed 1 : 1 during the organoid preparation. At each timepoint, duplicates of a pool of three organoids were disaggregated using mechanical and chemical (StemPro Accutase, Thermofisher) dissociation. To monitor the transfer by secretion, organoids prepared of only acceptor or donor cells were cultured in the same culture medium separated by a 1 µm filter. For FACS analysis, cells were passed through a cell strainer to separate cell aggregates and fixed in 2% PFA. Flow cytometry data were acquired with a BD Symphony A5 flow cytometer. GFP and mCherry fluorescence were analyzed at 488 nm and 561 nm excitation wavelengths, respectively. Ten thousand events were acquired for each condition and data were analyzed using FlowJo analysis software.
The extracellular flux cell mitochondrial stress analysis (Seahorse assay)
An extracellular flux analyzer (Seahorse XF96, Agilent, U.S.A.) was applied to analyze the mitochondrial function. The XF96 possesses a specialized microplate that allows for the measurement of the oxygen consumption rate (OCR) in real-time [36]. To test mitochondrial respiration, an XF Cell Mito Stress Test kit (Seahorse Bioscience; Agilent Technologies, Inc.) was used according to the manufacturer's protocol. The day previous to assay, the cells were seeded at a density of 20 000 cells/well in a laminin-precoated seahorse plate and incubated overnight at 37°C and 5% CO2. The sensor cartridge was hydrated in pure water at 37°C in a non-CO2 incubator overnight. On the day of the assay, the sensor cartridge was incubated in XF Calibrant 1 h at 37°C in a non-CO2 incubator prior to the assay. The culture medium was refreshed 1 h prior to the assays using unbuffered DMEM (pH 7.4) supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose (Seahorse Bioscience; Agilent Technologies, Inc.). Briefly, 1 µM oligomycin, 1 µM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), and 0.5 µM rotenone/antimycin A were subsequently added to the microplates. This enabled determination of the basal level of oxygen consumption, ATP-linked oxygen consumption, non-ATP-linked oxygen consumption, the maximal respiration capacity, and the non-mitochondrial oxygen consumption. A total of three basal OCR measurements were recorded prior to the injection of oligomycin. The decreased level of OCR represented oligomycin-sensitive OCR due to its inhibition of ATP synthase (complex V). FCCP, an uncoupling protein, was then injected and the FCCP-stimulated OCR was used to calculate spare respiratory capacity, which was defined as the difference between maximal respiration and basal respiration. The third injection was a mixture of rotenone (a complex I inhibitor) and antimycin A (a complex III inhibitor). This combination inhibited mitochondrial respiration completely, and thus no oxygen was further consumed by cytochrome c oxidase. The remaining OCR measurement obtained following this treatment was primarily non-mitochondrial and may have been due to cytosolic oxidase enzymes.
Irradiation
Irradiation was performed with X-Ray machine (Xstrahl LTD). 2 Gy irradiation was performed exposing the cells to X-rays for 1 min and 25 s (250 kV, 12 mA).
RT-qPCR
Total RNA extraction was performed using the RNeasy Mini Kit purchased from Qiagen. Reverse transcription was done using the Bio-Rad iScript gDNA Clear cDNA Synthesis Kit. Oligonucleotides were designed using Prime PCR Look Up Tool (Bio-Rad), purchased from Eurofins Genomics, and sequences are presented in Supplementary Table S1. Quantitative PCR was then performed using the Bio-Rad iTaq™ universal SYBR® Green supermix and analyzed using a CFX96TM real-time PCR detection system under the CFX Manager software (Bio-Rad). Gene expression was normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT), each point of each independent experiment was performed in triplicates. Oligonucleotides used in qPCR are presented in Supplementary Table S1.
Statistical analysis
The statistical tests for percentage of connected cells and percentage of transfer were computed using either a logistic regression model computed using the ‘glm’ function of R software (https://www.R-project.org/.) or a mixed effect logistic regression model using the lmer [37] and lmerTest [38] R packages. For cell connection in 2D, a mixed effect logistic regression model was estimated, adjusted on the effect of cell type, timepoint and condition. This model was also adjusted on the second and third order interactions among these three covariates. A random effect corresponding to replication of the experiment was also added to the model in order to account for potential batch effect. For percentage of transfer, we estimated a mixed effect logistic regression model adjusted on the condition, the day and the number of organoids. Second order interactions among condition and day and among number of organoids and day were added to the model in order to normalize statistical tests on time-varying heterogeneity of the number of organoids. A random effect corresponding to replication of the experiment was also added to the model in order to account for potential batch effect. All statistical tests to compare groups (among either cell lines, timepoints or treatments) were deduced by computing contrasts of one the abovementioned logistic model. P-values were therefore adjusted using Tukey's method. To compare the gene expression measured by RT-qPCR, Holm–Sidak method was applied to determine statistical significance, with alpha = 5. ANOVA two-way test was performed to compared cell number at different timepoints of the adherent culture. For the comparison of cell number in tumor organoids, the number of cells was transformed in logarithmic scale and slopes were compared.
Results
Patient-derived GBM cells with stem-like features form TNT-like connections
We obtained two GBM cells from a single tumor from a patient in the frame of the clinical trial STEMRI (Identifier: NCT01872221). This trial was aimed at studying the tumoral cells surrounding the core tumor after the latter has been surgically removed, and better understanding and possibly anticipating which of them are at the origin of the relapse. Two bulks of tissue were resected from the infiltrative tumor area defined by the Fluid-attenuated inversion recovery (FLAIR) sequence on MRI (Figure 1A) and characterized by multimodal MRI spectroscopy for their metabolic activity through the measurement of the Choline/N-AcetylAspartate Index (CNI) [39,40] (Supplementary Figure S1A). The tissues were then desegregated and cultured in stem cell medium [7] to enrich in GBM stem cells rather than differentiated ones [33], and grown in suspension in neurosphere-like aggregates (Figure 1B). To further characterize the two populations, named C1 and C2, we monitored the expression of genes associated with different cell types, from differentiated to progenitor/stem cells [4,6,41–44]. GFAP and CHI3L1 (respectively, astrocytic and mesenchymal markers) were not expressed and low expression was observed for the neural markers Tubβ3 and GAP43. On the other hand, expression of the progenitor and stem cell markers Olig1, Olig2, Sox11 and Sox2 was significant (Figure 1C). This pattern of gene expression was retained over passages in culture indicating maintenance of the stemness properties distinctive of GBM stem-like cells (GSLCs) [3]. We further characterized the C1 and C2 cells by Agilent Seahorse XF Cell Mito Stress Test allowing us to monitor mitochondrial function by directly measuring the aerobic respiration of cells. We observed that C2 and C1 cells behave in accordance to their respective region of origin (Supplementary Figure S1A). Indeed, the C2 population, derived from the highest metabolic area of the external tumoral region (CNI > 2), displayed higher maximal respiration and spare respiratory capacity compared with C1 cells originated from a lower metabolically active area (CNI < 2). On the contrary, C1 cells presented higher levels of non-mitochondrial oxygen consumption and basal respiratory capacity than C2 cells (n = 3, Supplementary Figure S1B). These data are consistent with a higher mitochondrial oxidative phosphorylation phenotype in C2 than in C1.
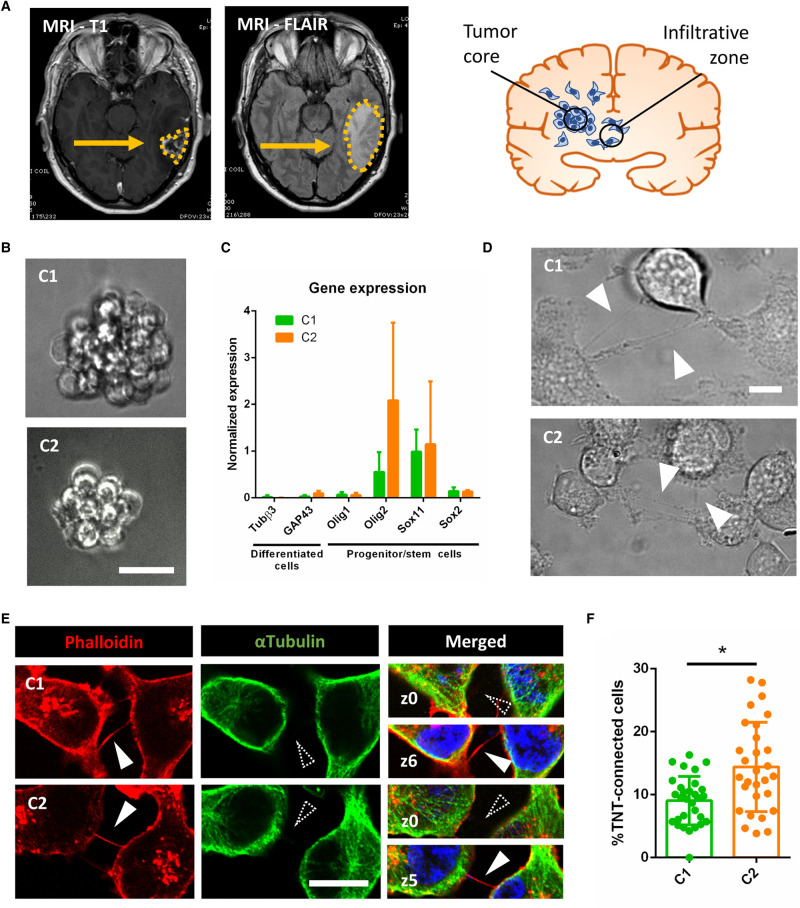
Figure 1. GSLCs form TNT-like structures.
(A) MRI analysis of C patient glioblastoma. The tumor is composed by a compact cellular part defined ‘Tumor core’, identified by MRI T1-Gadolinium (on the left). Some tumoral cells infiltrate the normal tissue, forming the ‘Infiltrative zone’ which is identified by MRI-FLAIR (right picture and schematics). C1 and C2 cells were obtained from different parts of the infiltrative zone (see Supplementary Figure S1). (B) C1 and C2 cell growth, forming neurosphere-like clusters in suspension. The resulting image represents a Z-projection of 30 and 50 slides (step size: 0.5 µm), respectively, acquired in Bright field using 40× magnification. Scale bar = 10 µm. (C) Expression of differentiation and progenitor/stem cells markers in C1 and C2, respectively in green and orange. The relative gene expressions were quantified by RT-qPCR after RNA extraction. Data were normalized over the expression of HPRT, housekeeping gene. GFAP and CHI3L1 showed no expression in both C1 and C2 and are not represented on the graph. The graph represents the means with SD of five independent experiments, each point performed in triplicate. P values >0.05 are not significant and not indicated on the figure. (D) GSLCs connected by TNT in live imaging in 2D culture. Cells were seeded on laminin-coated plates and pictures were taken after 6 h of seeding using 60× 1.4NA CSU oil immersion objective lens using Bright field. Arrowheads point to TNT-like connections. Scale bar = 20 µm. (E) GSLC TNTs containing actin but not microtubules. Cells were plated on laminin-coated surface for 6 h, fixed and stained with phalloidin (actin filaments, red), anti-αTubulin (microtubules, green) and DAPI (nuclei, blue). Representative images were acquired showing TNTs, actin-positive and αTubulin-devoid, floating above the dish surface. White-filled arrowhead indicates presence of TNT labeling, dashed arrowhead indicates absence of TNT staining. Scale bar = 10 µm. (F) Quantification of TNT-connected cells in C1 and C2, respectively in green and orange. GSLC were plated on laminin-coated surface, fixed after 6 h and stained with WGA. 2 × 2 tiles images were acquired with 60× objective and analyzed by Icy software. C1 were forming 9.0 ± 4% of connecting cells (five independent experiments each performed in duplicates, total n cells counted = 1239), while C2 were forming 14.4 ± 7% (four independent experiments, each performed in duplicates, total n cells counted = 1367), significantly more connected cells than C1 (P = 0.0370). Each dot represents an image containing an average of 40 cells each. P-values were deduced from contrast comparing the two cell populations in a logistic regression model. Error bar = standard deviation. P value <0.05 (*).
Different GBM-derived cell lines have been described to form TNT-like connections and to be able to transfer cellular content including mitochondria [28–31]. Because such cell lines are only partially recapitulative of the original tumoral features [45], here we aimed to address whether patient-derived GSLCs can form functional TNTs. Thus, C1 and C2 cells were plated for 6 h on laminin-coated surface in 2D culture to make them adhere to the support for ease of TNT recognition. Thin cell connections were detected after 6 h of culture by live imaging (Figure 1D). After fixation, we assessed the presence of actin-containing connections floating above the laminin-coated surface, which is a distinguishing characteristic of TNTs, that hoover above the substrate, as exemplified in Figure 1E, where both attached-to-the-substrate (z-stack = 0) and above (z-stack > 3) stacks are shown [35,46]. GSLC TNTs resulted to be always positive for actin and negative for microtubules markers, consistent with the description of classical TNTs [16] (Figure 1E). For quantification of TNTs, we labeled the cell's plasma membrane with fluorescent-Wheat Germ Agglutinin (WGA) and counted thin, continuous, non-attached to the substratum protrusions [35] connecting distant cells. Both C1 and C2 populations formed TNT-like connections with a significantly different frequency: ∼10% of TNT-connected cells in C1 and 15% in C2 (Figure 1F). These data showed for the first time that GSLCs, derived from the infiltrative region of the tumor, can interconnect through TNT-like structures.
TNT-like structures of GSLCs can transfer mitochondria
TNTs are described to be open-ended connections allowing the passage of cellular cargoes. To determine whether the connections observed were apt to this purpose, we decided to assess the transfer of mitochondria, shown to occur in several types of cancer cells (mesothelioma, leukemias, ovarian, etc.) [13,14,27]. To observe mitochondria in living samples, we introduced a GFP-tagged fragment of the subunit VIII of human cytochrome C oxidase located in the inner mitochondrial membrane (MitoGFP) in both GSLCs by lentiviral transduction. We then performed live-imaging on GSLCs and found mitochondria moving inside a TNT-like structure and entering into a connected cell (Figure 2A, Supplementary Video S1), supporting an open-ended TNT relying the two cells. To quantify this transfer, we performed co-culture assays [35] between a donor population, expressing MitoGFP, and an acceptor cell population transduced with lentivirus governing the expression of cytosolic mCherry (Figure 2B). More than 80% of each cell population was stably expressing the constructs, allowing a 1:1 co-culture ratio between the two populations. Cells were plated on a laminin-coated surface and grown either in direct contact or separated by a filter (transwell) (Figure 2B), and the percentage of mitochondria transfer was assessed after 2 or 5 days of co-culture by flow cytometry [35]. Between 1 and 3% of acceptor cells received donor-derived mitochondria, exclusively due to a contact-dependent mechanism since negligible transfer was observed when the two cell populations were separated by filter but shared the same medium (Figure 2C). Furthermore, the percentage of acceptor cells receiving mitochondria was increasing over time in both GSLCs (Figure 2D). It is worth noting that C2 had higher mitochondria transfer compared with C1, in accordance with the higher percentage of TNT-connected cells in this population (Figure 1F). As both GSLCs had a similar proliferation rate in this condition (Figure 2E) we could rule out that the different transfer abilities of the two cell types were related to differences in cell densities. Next, to confirm that the fluorescence signal detected by flow cytometry corresponded to true mitochondria inside acceptor cells, confocal microscopy was performed in the same co-culture conditions used for the flow cytometry experiments. By this mean MitoGFP puncta (Figure 2F) which overlapped with TOM20 (Translocase of the Outer Membrane, Supplementary Figure S2) were observed in acceptor cells. These data indicate that both C1 and C2 GSLCs are able to form functional TNTs when cultured in 2D. However, C1 and C2 transfer mitochondria with distinct efficiencies, consistent with their distinct abilities to form TNTs (Figure 1F).
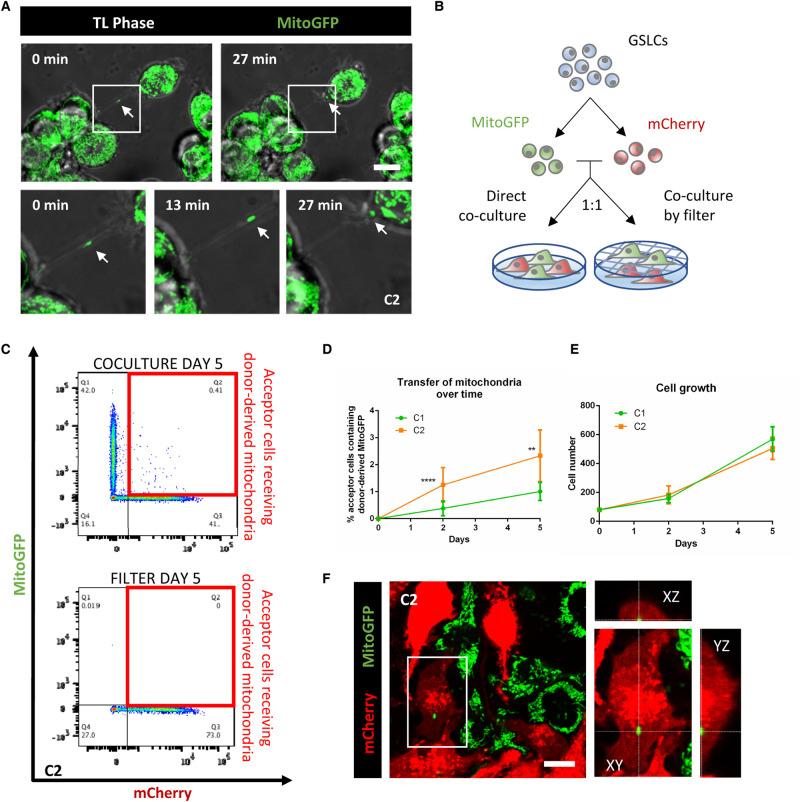
Figure 2. GSLCs transfer mitochondria through TNTs.
(A) C2 expressing MitoGFP are connected by TNTs containing mitochondria. Cells were seeded on laminin-coated dish and after 6 h video were acquired using Bright field and laser 488 in a Spinning Disk microscope. Timeframes show the mitochondria moving along the connection and entering in one of the two connected cells. Each timeframe of the video is the result of the Z-projection of 18 slides (step size: 0.5 µm) (B) Schematic representation of the co-culture experiment. Donor MitoGFP cells were co-cultured with acceptor mCherry cells at 1 : 1 ratio either by direct contact or through a 1 µm filter. (C) Representative flow cytometry plot of C2 after 5 days of co-culture. Acceptor and donor cells, respectively lie on the X and Y axis. Acceptor cells positive for MitoGFP signal are framed in the red boxes. (D) Quantification by flow cytometry of the mitochondria transfer over time in C1 and C2, respectively in green and orange. A minimum of 10 000 events were analyzed after 2 or 5 days of co-culture. C1 shows 0.38 ± 0.27% and 1.01 ± 0.33% of acceptor cells receiving mitochondria after 2 and 5 days, respectively (four independent experiments, each performed in duplicate). C2 shows 1.25 ± 0.63% and 2.33 ± 0.95% of acceptor cells receiving mitochondria after 2 and 5 days, respectively (five independent experiments, each performed in duplicate), significantly more than C1 (P < 0.0001 (****) at day 2, P = 0.0085 (**) at day 5). P-values were deduced from contrast comparing the two cell populations in a logistic regression model. Error bar = SD (E) Cell growth in co-culture experiment. 80 000 GSLCs per well were plated at time 0 and counted after co-culture. For C1, 158 000 ± 28 751 and 568 866 ± 85 332 cells were counted after 2 and 5 days, respectively (three independent experiments). For C2, 182 900 ± 61 890 and 505 260 ± 77 515 cells were counted after 2 and 5 days, respectively (five independent experiments). Error bar = SD. ANOVA two-way test was performed and showed no significant difference between C1 and C2 at the two timepoint in analyze. (F) Representative image of co-culture assay in C2. Donor MitoGFP (in green) and acceptor mCherry cells (in red) were fixed after 5 days of co-culture, confocal images were acquired with 63× objective. In the magnification, the orthogonal view of an acceptor cell containing donor-derived mitochondria. Scale bar = 10 µm.
TNT-like structures exist in GSLC tumor organoids together with TM-like protrusions
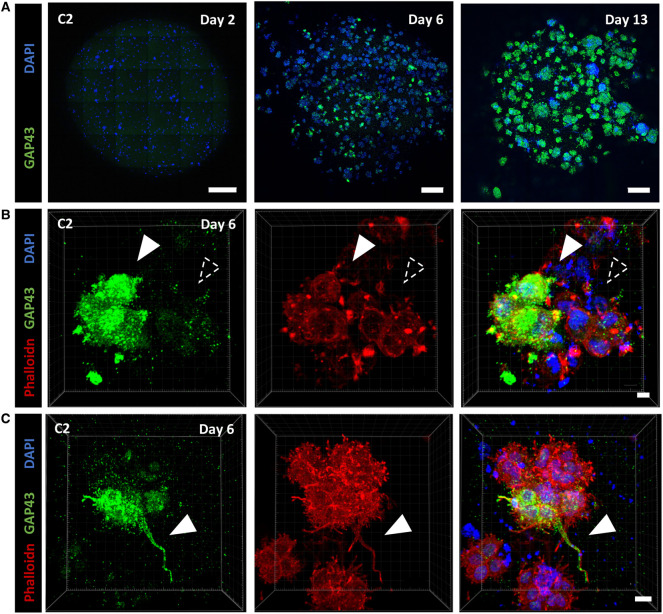
To assess the participation of the TNT-mediated communication to GBM networking in a context more representative of the tumor, we cultured individually each GSLC in tumor organoids, according to the protocol published by J. Rich and colleagues [34]. Tumor organoids are a relevant, 3-dimensional, culture method which allows long-term growth preserving the stem cell identity of some cells and reconstitutes, to some extent, the morphological and phenotypic heterogeneity of the original tumor [34]. We were able to grow C1 and C2-derived tumor organoids up to more than 23 days of culture. To characterize the transcriptional changes undergone by the cells in this culture system, we quantified the expression of differentiation and progenitor/stem markers in 23-days-old organoids by RT-qPCR. We observed no significant variation in the expression of all tested genes compared with classical culture, except of GAP43 in C2 cells (Figure 3B), that resulted to be 12-fold higher in organoids. This result showed that although most of the cells still expressed progenitor markers, maintaining therefore their multipotency, some could also commit to a more differentiated profile, as it happens in vivo.
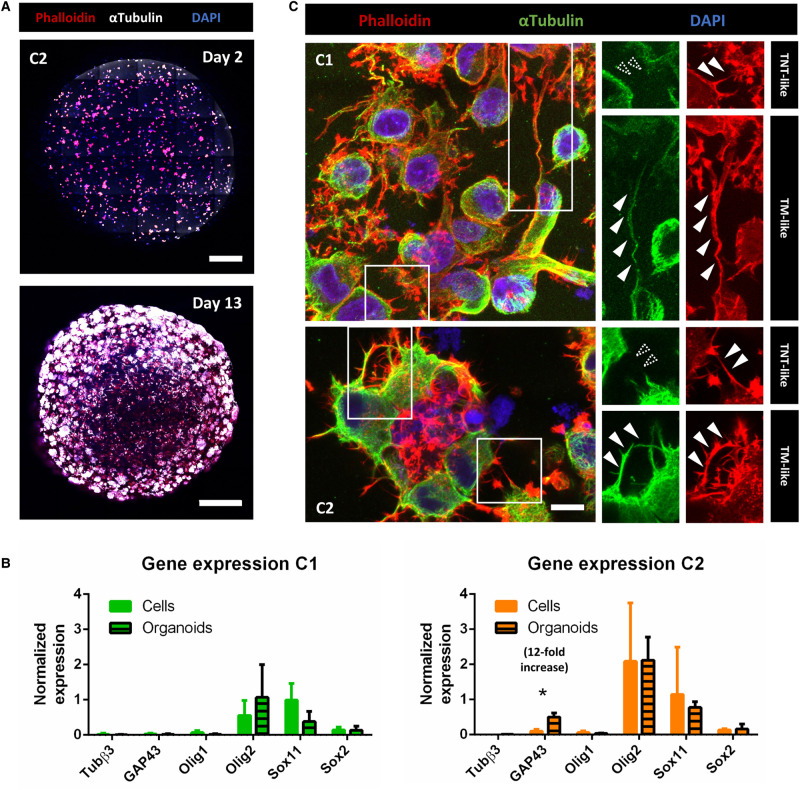
Figure 3. GSLCs in tumor organoids.
(A) Images of C2 tumor organoids at 2 and 13 days of growth using Pln-Apo 10×/0.45 objective of inverted confocal LSM700. The resulting images represent a max intensity projection of 5 and 31 sections (step size: 7 and 3.12 µm), respectively, stained for anti-αTubulin (microtubules, white), Phalloidin (actin in red) and nuclei (blue). Scale bars are 200 (top) and 500 µm (bottom). (B) Expression of differentiation and progenitor/stem cells markers in C1 and C2 organoids, respectively in green and orange. The relative gene expressions were quantified by RT-qPCR after RNA extraction from 23-days-old organoids, normalized over the expression of HPRT. Note the 12-fold increased expression of GAP43 in C2 tumor organoids, and GFAP and CHI3L1 show no expression in both conditions and are not represented on the graph. The graphs represent means with SD of three and four independent experiments for C1 and C2 respectively, each point performed in triplicate. Holm–Sidak method was applied to determine statistical significance between cells and organoids for each gene. P value <0.05 (*), P values >0.05 are not significant and not indicated on the figure. (C) C1 and C2 tumor organoids at 9 and 6 days, respectively, stained for anti-αTubulin (microtubules, green), Phalloidin (actin filaments, red), and nuclei (blue). Confocal images were acquired with 40× objective. Regions of interest show either αTubulin-devoid connections, defined as TNT-like (<1 µm), or thick αTubulin-positive connections (>1 µm), named TM-like. Dashed arrowheads indicate absence of fluorescent signal at the connection level, white-filled arrowhead show positiveness to the signal. Both images are max intensity projections of 12 slices (step size: 0.38 µm). Scale bar = 10 µm.
To date, TNT visualization in 3D cultures and their quantification had not been reported, as preserving and identifying these fragile thin structures in 3D is extremely challenging. In both C1 and C2-derived tumor organoids, fluorescently labeled with phalloidin and anti-αTubulin antibody and imaged with confocal microscopy, we observed different types of cell protrusions (Figure 3C). Specifically, we noticed thin (<1 µm), actin-rich structures (devoid of tubulin) resembling TNTs that were found connecting cells already within the first week of organoids culture. However, the resolution of light microscopy does not allow to assess if these connections were functional TNTs or close-ended protrusions (like filopodia) [16]. One possibility to address this, is to look for the presence of organelles inside the connections and assess transfer. To this aim, first we imaged tumor organoids prepared with GSLCs stably expressing MitoGFP construct. Several cell extensions were found to be rich in content of mitochondria (Supplementary Video S2). Of note, by using live-imaging we observed thin TNT-like connections containing mitochondria trafficking between two connected cells (Figure 4A, Supplementary Video S2, white arrow, and Supplementary Figure S3), in accordance with what was observed in 2D. Next, to quantify mitochondrial transfer, we prepared tumor organoids mixing MitoGFP donor and mCherry acceptor cells in a 1 : 1 ratio. After 6, 9, 13, 16, 20 and 23 days of co-culture inside the same organoids, duplicates of three organoids per condition were desegregated (and combined) in a single cell suspension and analyzed by flow cytometry for the presence of MitoGFP into acceptor mCherry-positive population (Supplementary Figure S5). The percentage of acceptor cells receiving mitochondria was increasing over time, reaching ∼3% in C1 and 8% in C2 after 23 days of culture (Figure 4B, note the logarithmic y axis scale). Higher efficiency of transfer was observed in C2 cells when comparing the general trend of the transfer with the one of C1 (Figure 4B), in agreement with the data obtained when cells were cultured in 2D. Mitochondria transfer was not related to cell proliferation as both GSLCs grow similarly in organoids (Figure 4C). Overall, these data were consistent with the results obtained in 2D and suggested that C2 have higher ability to form and use TNTs for transferring cellular content, compared with C1. To verify that mitochondria transfer was not due to secretion, we co-cultured organoids composed of only one cell population, either donor or acceptor cells, in the same medium. We have not observed any transfer of MitoGFP from donor organoids to acceptor organoids in these conditions over time (Supplementary Figure S5), strongly suggesting that the mitochondria transfer that we quantified in mixed organoids was dependent on direct cell contacts between donor and acceptor cells. Overall, these data also indicate that GSLCs form functional TNT-like connections able to transfer mitochondria.
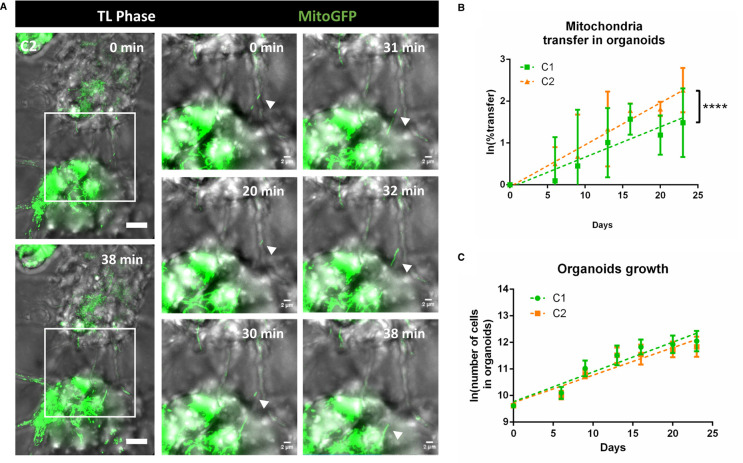
Figure 4. Mitochondria transfer in tumor organoids.
(A) TNT-like connection between C2 cells containing mitochondria in 6-days old tumor organoids. Timeframes result of the max projection of 62 slides (step size: 0.5 µm) with a total physical thickness of 31 µm, with 1 minute of interval time. White arrows point to the mitochondria movement inside the TNT at the different time points. Video were acquired using Bright field and laser 488 in a Spinning Disk microscope. (B) Quantification of the mitochondria transfer in tumor organoids over time in C1 and C2, respectively in green and orange. Organoids were prepared mixing donor and acceptor cells for each GSLC. For each timepoint and condition, duplicates of a pool of three organoids were dissociated in a single cell suspension and fixed for flow cytometry analysis after 6, 9, 13, 16, 20 and 23 days of culture. All the cells in the suspension were analyzed to obtain the percentage of acceptor cells receiving mitochondria. C1: day 6 1.54 ± 1.4%; day 9 2.80 ± 2.9%; day 13 2.20 ± 1.1%; day 16 5.07 ± 2.06%; day 20 3.55 ± 1.5%; day 23 3.05 ± 0.84% (four independent experiments). C2: day 6 1.72 ± 0.7%; day 9 2.64 ± 2.2%; day 13 4.96 ± 4.35%; day 16 5.98 ± 1.02%; day 20 5.57 ± 0.03%; day 23 8.37 ± 2.7% (three independent experiments). Percentage of transfer was transformed into a logarithmic scale. Error bar = SD. P-values are deduced by comparing the slopes of the two cellular population in a logistic regression model as described in material and methods. P value <0.0001 (****) (C) Cell number in tumor organoids. For each timepoint and condition, duplicates of a pool of three organoids were dissociated in a single cell suspension C1: day 6 24 800 ± 5768; day 9 63 150 ± 18 350; day 13 105 850 ± 43 970; day 16 140 450 ± 33 929; day 20 158 600 ± 60 394 day 23 181 800 ± 78 820 (four independent experiments). C2: day 6 22 600 ± 3704; day 9 49 700 ± 8116; day 13 104 200 ± 33 870; day 16 108 580 ± 42 218; day 20 128 800 ± 34 478; day 23 145 080 ± 47 726 (four independent experiments). The cell number was transformed into a logarithmic scale and slopes were compared by linear regression (dashed lines). No significant difference was observed between C1 and C2.
Interestingly, in addition to the TNT-like connections described above, we also observed thick (>1 µm) and long protrusions containing both actin and microtubules rather similar to TMs (previously observed in vivo [10], but not in the 2D cultures), which coexisted with TNTs in the same organoids (Figure 3C). To better characterize TM-like protrusions we decided to evaluate the presence of GAP43, a TM-marker described to be one of the major driver of TM formation [10]. Intriguingly, by RT-qPCR we had observed increased expression of this marker in C2- but not in C1-derived tumor organoids (Figure 3B). In agreement with these data, by immunofluorescence we observed an increase in the number of GAP43-positive C2 cells over time (see days 2, 6 and 13 labeling's in Figure 5A,B), confirming that tumor organoids reproduced to some extent tumoral heterogeneity and structure. In addition, part, but not all, of TM-like extensions resulted positive for GAP43 (Figure 5C) in C2 organoids. Of interest, some of these thick TM-like structures in C1 and C2 organoids contained mitochondria (Supplementary Video S2, red arrow). This was consistent with the observations of Winkler and colleagues [10], and also in our case mitochondria transfer was not detected through such structures [47]. The presence of these two types of connections, TM-like (also expressing GAP43) and TNT-like, was further confirmed in organoids produced from one GSLCs (O) derived from a second patient (Supplementary Figure S4), suggesting that the coexistence of TNTs and TMs in GBM could be recurrent in different tumors. Taken together, these results showed that the 3D organoid model using GSLCs is a valid representation of the tumoral complexity in vivo [34] and that various types of connections, including TNTs and TMs, could coexist in the network formed by GSLCs. However, only TNTs could provide a route for the exchange of cellular material, notably mitochondria.
Figure 5. GAP43 expression and TM characterization in tumor organoids of GSLC cells.
(A) GAP43 protein expression increases over time. 2, 6 and 13 days-old organoids were fixed and stained with anti-GAP43 (in green) and DAPI (in blue). Confocal images with 10× objective were acquired. Images result from the max intensity projection of 5, 20, 11 sections (step size: 7, 3.13, 3.13 µm), respectively. Scale bars: 500 µm, 200 µm, 200 µm (from left to right). (B) Heterogeneous expression of GAP43 in C2 tumor organoids. 6 days-old C2 organoids were fixed and stained with anti-GAP43 (in green), phalloidin (actin filaments, in red) and DAPI (in blue). Confocal images with 63× objective were acquired. 3D reconstruction of a 50-sections image (step size: 0.33 µm) was performed using Imaris Viewer software. White-filled arrowhead point to a cluster of cells expressing GAP43, alternatively a group of cells negative for its expression are indicated with a dashed arrowhead. Scale bar: 5 µm. (C) TM-like protrusion can express GAP43 in C2 organoids. Six days-old C2 organoids were fixed and stained with anti-GAP43 (in green), phalloidin (actin filaments, in red) and DAPI (in blue). Confocal images with 63× objective were acquired. 3D reconstruction of a 77-sections image (step size: 0.77 µm) was perfomed using Imaris Viewer software. White-filled arrowheads point toward a TM-like extension expressing GAP43. Scale bar: 15 µm. 3D reconstructions were performed with Imaris Software.
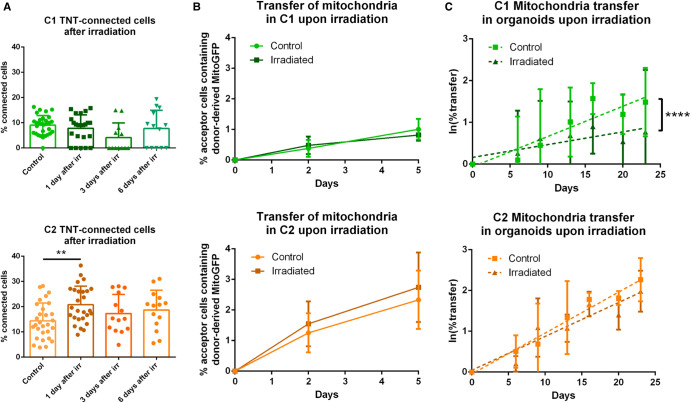
Effect of irradiation on TNT-based communication in C1 and C2 GSLCs
GBM relapse, originates from GSLCs [4] and treatments introduce alterations in GBM cells that seem to favor their survival [7]. As TNTs have been involved in cancer progression of different tumors [13] we decided to assess the effect of irradiation on the TNT-based communication in GSLCs. We irradiated cells at a dose of 2 Gray (Gy). This dose, daily administrated to GBM patients for 5 days a week for 6 weeks during radiotherapy [1], was applied only once on our cells in order to be effective but subtoxic [7] and preserve cell viability for all the duration of the experiment. Cells were plated on laminin-coated coverslips for 6 h at 1, 3 and 6 days after the irradiation, and next were fixed and analyzed for their TNT content. While C1 cells showed a slight decrease, not statistically significant, in their percentage of TNT-connected cells after irradiation, TNT frequency was significantly increased in C2 cells the day following the irradiation (Figure 6A), suggesting an acute effect induced by the irradiation in this population. To assess the effect of irradiation on the transfer of mitochondria, 2 Gy irradiation was applied on the donor cells the day before the co-culture and transfer was quantified after 2 or 5 days of co-culture. The percentage of C1 acceptor cells containing donor-derived mitochondria was not affected by irradiation, whereas we observed a tendency to an increased transfer upon irradiation in C2 (Figure 6B), although not statistically significant. Of note, in this experiment, C1 cells showed mild but significant reduction in their cell number at 5 days from irradiation, compared the control condition, differently from C2 cells which did not show significative variations (Supplementary Figure S6A). To assess variations of mitochondria, transfer upon irradiation on a long-term co-culture we irradiated C1 and C2 -derived organoids at day 5 of culture and assessed mitochondria transfer at various timepoints. After 23 days in culture, 3% and 1.7% of transfer was observed, respectively, in C1 control and irradiated condition, showing a significant decrease in transfer in the irradiated condition (Figure 5C). On the other hand, in C2 there was no significant reduction in the transfer efficiency over time, with 8% and 7% of transfer observed in control and irradiated condition, respectively (Figure 6C). The different behavior of C1 and C2 cells was independent of the effect of irradiation on cell growth as the number of cells into both types of organoids was not significantly affected by irradiation (Supplementary Figure S6B), as expected with the chosen doses. Importantly, when co-culturing single population organoids in the same dish to assess the contribution of secretion in transfer we did not observe transfer.
Figure 6. Effect of GSLC irradiation on TNT-based communication.
(A) Quantification of TNT- connected cells in C1 and C2 after irradiation, respectively in green and orange, in adherent cell culture. GSLCs were irradiated 1 day before cell plating on laminin-coated surface, then fixed after 6 h and stained with WGA. 2 × 2 tiles images were acquired with 60× objective and analyzed by Icy software, experimental duplicates were performed for each condition. The graphs represent means with SD. C1 were forming 7.8 ± 5% (four independent experiments, tot n cells counted = 891), 4.1 ± 6% (three independent experiments, total n cells counted = 300) and 7.7 ± 7% (three independent experiments, total n cells counted = 313) of connecting cells after 1, 3 and 6 days from the irradiation, respectively. No statistically significant difference was observed compared with control (9.0 ± 4%, five independent experiments, total n cells counted = 1239). C2 were forming 20.8 ± 7% (four independent experiments, total n cells counted = 1368), 17.3 ± 7% (three independent experiments, total n cell counted = 552) and 18.7 ± 8% (three independent experiments, total n cells counted = 462) of connecting cells after 1, 3 and 6 days from the irradiation, respectively. A statistically significant increase was observed 1 day after irradiation compared with control (14.4 ± 7%, four independent experiments, total n cells counted = 1367, P = 0.0073 (**)). Each dot represents an image containing an average of 40 cells each. P-values were deduced from contrast comparing the two cell populations in a logistic regression model. (B) Quantification of the mitochondria transfer by flow cytometry in both GSLCs over time in C1 and C2 upon irradiation, respectively in green and orange. Donor GSLC were irradiated 1 day before the co-culture, analysis was performed after 2 or 5 days (corresponding at 3 and 6 days from the irradiation). A minimum of 10 000 events were analyzed per condition, each performed in duplicate. In irradiated condition, C1 show 0.48 ± 0.28% and 0.81 ± 0.18% of acceptor cells receiving mitochondria after 2 and 5 days, respectively (four independent experiments). No statistically significant difference was observed compared with control (day 2: 0.38 ± 0.27%; day 5 1.01 ± 0.33%; four independent experiments). In irradiated condition, C2 show 1.54 ± 0.73% and 2.74 ± 1.13% of acceptor cells receiving mitochondria after 2 and 5 days, respectively (five independent experiments). No statistically significant difference was observed compared with control (day 2: 1.25 ± 0.63%; day 5: 2.33 ± 0.95%. Five independent experiments). P-values were deduced from contrast comparing the two cell populations in a logistic regression model. Graphs are means with SD. (C) Quantification of the mitochondria transfer in tumor organoids upon irradiation in C1 and C2, respectively in green and orange. Organoids were prepared mixing donor and acceptor cells for each GSLC and irradiated at 5 days from their preparation. Experiment was performed as in Figure 4D. Control C1: day 6 1.54 ± 1.4%; day 9 2.80 ± 2.9%; day 13 2.20 ± 1.1%; day 16 5.07 ± 2.06%; day 20 3.55 ± 1.5%; day 23 3.05 ± 0.84%. Irradiated C1: day 6 1.90 ± 1.6%; day 9 4.45 ± 1.9%; day 13 2.50 ± 1.7%; day 16 2.82 ± 1.5%; day 20 2.39 ± 1.61%; day 23 1.76 ± 1.2% (four independent experiments, P < 0.0001, ****). Control C2: day 6 1.72 ± 0.7%; day 9 2.64 ± 2.2%; day 13 4.96 ± 4.35%; day 16 5.98 ± 1.02%; day 20 5.57 ± 0.03%; day 23 8.37 ± 2.7%. Irradiated C2: day 6 1.23 ± 0.2%; day 9 3.50 ± 2.3%; day 13 3.03 ± 0.9%; day 16 5.46 ± 1.5%; day 20 4.23 ± 1.3%; day 23 7.21 ± 1.7% (three independent experiments, P = 0.0665). Percentage of transfer was transformed into a logarithmic scale. P-values are deduced by comparing the slopes of the two cellular population in a logistic regression model as described in material and methods. Error bar = SD.
Overall, consistent with the wide heterogeneity present in GBM tumors where distinct molecular profiles coexist and exhibit differential therapeutic responses [3], our data indicated that GSLCs derived from the same tumor display intrinsically different properties and have diverse responses to irradiation regarding the effect on TNT formation and transfer function. Specifically, TNT functionality was preserved in C2 organoids after irradiation whereas it was reduced in C1 organoids.
Discussion
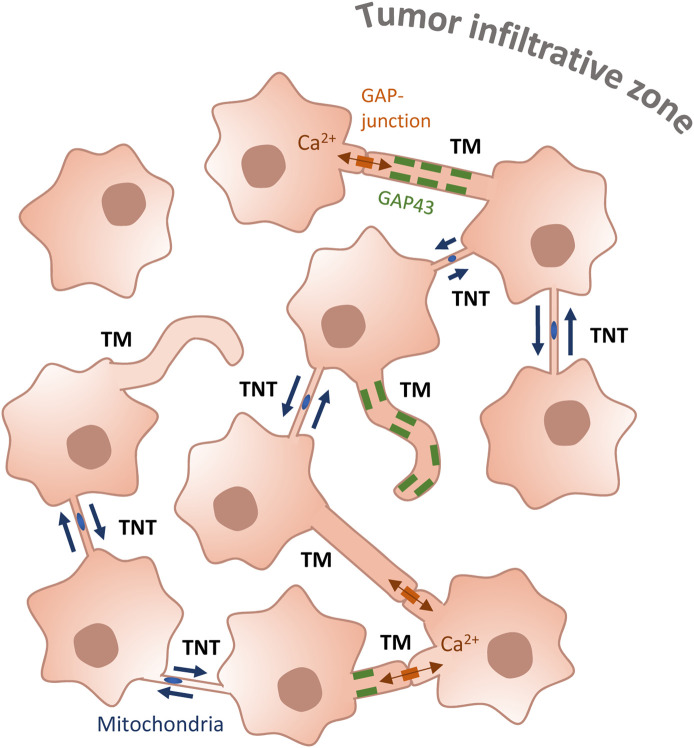
Tunneling nanotubes (TNTs) are gaining an increasing relevance in the context of cancer development and progression [13]. Their presence and ability to transfer cellular material including organelles, has been correlated with the induction of migratory ability, angiogenesis, cell proliferation ad therapy-resistance [14,27]. Few reports have addressed the presence of TNTs in GBM, where it was shown that GBM-derived cell lines in culture were able to form TNT-like connections [28,29,32] and exchange mitochondria with healthy cells of the tumor microenvironment, while promoting cell aggressive phenotypes as increasing proliferation and treatment-resistance [30,31,48]. Nevertheless, at transcriptomic level GBM cells lines are drastically divergent from the original tumor [45], thus they do not appear to be a very relevant model for GBM. On the other hand, TNTs were not reported in patient-derived GSCs xenografted into murine brains, which formed a complex tumor cell network based on thicker neurite-like connections, called TMs [10]. In this elegant work the authors used live two-photon microscopy to image the implanted tumor with a resolution that might have not been sufficient to visualize TNTs. Thus, whether TNTs participate to this networking in a tumoral relevant model, but were not detected because of the approach used, is not yet clear. To address this question, here we have assessed their presence and functionality in two GSLCs derived from the most external tumoral area, responsible for GBM relapse. By using live-imaging and quantification of transferred mitochondria, we showed for the first time that GSLCs are able to interconnect and transfer mitochondria via TNT- like structures in 2D culture, in both GSLCs populations. Importantly, to visualize and characterize these connections and to monitor their ability to transfer mitochondria in a context closer to the tumor, we cultured these GSLCs as tumor organoids, a very relevant model shown before to recapitulate at phenotypic and transcriptomic level patient tumors heterogeneity [34,45,49]. Indeed, in our organoid cultures, GSLCs maintained their progenitor characters over more than 23 days. Using confocal microscopy in fixed conditions, we provided evidences for the existence of thin TNT-like protrusions connecting different cells in the organoids from early time in culture; while live- imaging demonstrated mitochondria moving along TNTs and entering into the connected cells. These observations supported the hypothesis that TNTs exist and are functional in GBM-derived tumor organoids, suggesting that TNT-based communication may be relevant in the actual tumor. Because TNT communicative abilities were observed in the GSLCs obtained from the infiltrative and putative relapse-driving area, we could speculate that this might have a potentially relevant role in tumor recurrence and induction of aggressive phenotypes, as shown in the case of other tumors [13,14]. Nevertheless, the aggressiveness of GBMs has been correlated before with the establishment of tumor networking based on TMs [10]. TMs are neurite-like extensions [50], able to connect to distant cells and propagate action potential [51,52] and proposed to scaffold the network formed by cancer cells. Interestingly, we also observed thicker connections resembling TMs in our GSLC tumor organoids. Although TMs are organelle-rich structures, no cargo transfer through them has been demonstrated [47], while the presence of GAP-junctions along their length [53] support their role in electric signal transmission. Our data confirmed these observations in organoids, since we could follow movements of mitochondria in cell bodies and protrusions, including TM-like structures. However, the effective transfer of mitochondria until entering a connected cell could be observed only through TNT-like connections, and not in TM [10,11,51]. Of note, we observed and quantified mitochondria transfer in both C1 and C2 GSLCs, grown in 2D culture conditions where none of them expressed GAP43, which is a typical marker and the major known driver of TMs [10,11], and in 3D organoids where only C2 subpopulations expressed GAP43, suggesting that the transfer of mitochondria does not occur through GAP43-dependent structures. Notwithstanding, we observed that C2 cells, which can express GAP43, were transferring mitochondria more efficiently compared with C1 cells, even after irradiation. These results are in accordance with the work of F. Winkler and collaborators [10,11] suggesting that GAP43 expression and the presence of TMs could be correlated to the aggressiveness of the tumor. Because we observed TNTs and TMs in the same organoid, we propose that TNTs and TMs coexist and co-operate in GBM networking, carrying on complementary roles that could participate eventually to treatment-resistance. In particular, we hypothesize that in a situation in which a TM tumor network is formed based on signaling exchanges between interconnected cells there will be the possibility to form more TNTs, which will provide the transfer function and material exchange (Figure 7). This hypothesis needs to be further tested in vivo, ideally in the conditions in which TMs have been observed before [10].
Figure 7. GSLC network.
GSLCs interconnect through different types of cellular extensions. TMs are thick (>1 µm) protrusions that can either contact other cells through GAP-junctions, allowing the propagation of calcium flux, or be individual finger-like extensions not connecting remote cells. They can be positive for GAP43 (rectangles along the membranes of TM), neuronal Growth-Associated Protein. GSLCs also interconnect through TNTs, thinner (<1 µm), open-ended connections which allow transfer of cellular cargos, such as mitochondria (ovals in TNTs).
In our GSLC model we have chosen to specifically look at mitochondria transfer; as mitochondria can provide metabolic support to cancer cells [14,27], transfer of mitochondria has been shown to modulate the response to treatments in a beneficial manner for the recipient cells impacting on their metabolism, rescuing their aerobic respiration and providing a metabolic support against treatment-related stress [27,54]. However the observation of mitochondria transfer between GSLCs does not preclude the possibility that other cellular material (e.g. RNA, proteins, other vesicles) could be additionally transferred through the same connections, as observed in other cancer types [55–57]. Because of the limitation of our study, it will be of important in the future to address first what is the exact composition of transferred material, and second what are the functional consequences of such events for both donor and recipient cells. Wide cellular heterogeneity is one of the many reasons that makes GBM very difficult to treat, as it is formed by cells with different abilities to respond to the treatments. We have shown that two GSLCs derived from two areas of the same tumor, also display different behavior, including percentage of TNT-connected cells and transfer of mitochondria over time in response to irradiation. Specifically, C2 cells, coming from the most metabolically active area of the tumor (CNI+) previously shown to be more likely leading tumor relapse [58], in our hands also displayed higher ability to respond to an energetic demand (maximal respiration and spare respiratory capacity, Supplementary Figure S1C) compared with C1 cells. It is conceivable that the presence of metabolic heterogeneity may offer to GBM tumor cells adaptive mechanisms to better respond and overcome the cellular stress introduced by the treatments [59]. As C2 cells were shown to maintain their TNT-communicative ability after irradiation, differently from C1 cells, it is tempting to speculate that TNTs might contribute to this rescue process. However, the potential link between metabolism and ability to grow TNTs and to respond to treatment needs to be directly investigated by functional studies and in a large number of patient-derived cells. Furthermore, our data do not exclude that other mechanisms (including release of various extracellular factors as well as TMs) are involved in GBM therapy-resistance. TNT-mediated communication has been described to be an advantageous feature for tumoral cells in several cancer models [13], the ability to exploit this way of communication could be common in more aggressive cancer cells, also in the case of GBM. Overall, our data are consistent with tumor networking being important in GBM progression, resistance to treatment and relapse. In addition to TMs, the ability to grow functional TNTs could participate to the formation of a functional tumor network, where exchanges of organelles and different material in addition to signaling molecules is allowed (Figure 7). Although our data represent a step up to the use of GBM derived cell lines, it will be necessary to further investigate their relevance in a larger sample of tumors to specifically analyze the impact of TNT-based networking in the context of tumor progression, therapy-resistance and relapse. In this context, it may as well be possible to exploit them by considering inhibition of TNT-dependent transfer as a new way for optimization of radiotherapy efficacy in GBM.
Conclusions
Our data suggest that TNT-mediated exchange of cellular material occurs between GSCs and that in addition to TMs, TNTs participate to GBM tumoral networking providing a route for the transfer of intracellular material and potentially contributing to tumor progression and treatment-resistance.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
We thank all the members of the MOGLIMAGING network, HTE program, Aviesan and INSERM which participated in our collaboration. We thank I. Leroux for teaching tumor organoids preparation and culture. We thank all the members of the UTRAF unit for their support.
Abbreviations
- CHI3L3
chitinase 3-like-3
- FCCP
carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- GAP43
growth-associated protein 43
- GBM
glioblastoma
- GFAP
glial fibrillary acidic protein
- GSCs
glioblastoma stem cells
- GSLCs
glioblastoma stem-like cells
- OCR
oxygen consumption rate
- Olig1
oligodendrocyte transcription factor 1
- Olig2
oligodendrocyte transcription factor 2
- Sox11
sex-determining-region-Y-Box transcription factor 11
- Sox2
sex-determining-region-Y-Box transcription factor 2
- TMs
tumor microtube
- TNTs
tunneling nanotube
- TOM20
translocase of outer membrane
- Tubβ3
class III β-tubulin
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was funded by grants from HTE (HTE201502) to CZ and EMJC, Institut National du Cancer (PLBIO18-103) to CZ and EMJC, Fondation de France (Fondation de France n: 00100228) to ISSM and Fondation ARC pour la recherche sur le cancer to GP (DOC20190508549).
Ethical Approval
The consent for the use of human material has been given with the clinical trial STEMRI (Identifier: NCT01872221).
CRediT Contribution
Chiara Zurzolo: Conceptualization, Resources, Supervision, Funding acquisition, Validation, Investigation, Project administration, Writing — review and editing. Giulia Pinto: Data curation, Formal analysis, Funding acquisition, Investigation, Visualization, Methodology, Writing — original draft. Inés Saenz de Santa Maria: Data curation, Formal analysis, Funding acquisition, Investigation, Visualization, Methodology, Writing — original draft. Patricia Chastagner: Data curation, Formal analysis, Investigation, Methodology. Emeline Perthame: Data curation, Software, Formal analysis. Caroline Delmas: Resources, Methodology. Christine Toulas: Conceptualization, Resources. Elizabeth Moyal-Jonathan-Cohen: Conceptualization, Resources, Funding acquisition, Validation, Writing — review and editing. Christel Brou: Conceptualization, Resources, Data curation, Formal analysis, Supervision, Validation, Investigation, Methodology, Writing — review and editing.
Supplementary Material
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2.Batash R., Asna N., Schaffer P., Francis N. and Schaffer M. (2017) Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr. Med. Chem. 24, 3002–3009 10.2174/0929867324666170516123206 [DOI] [PubMed] [Google Scholar]
- 3.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L.L. and Rich J.N. (2015) Cancer stem cells in glioblastoma. Genes Dev. 29, 1203–1217 10.1101/gad.261982.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prager B.C., Bhargava S., Mahadev V., Hubert C.G. and Rich J.N. (2020) Glioblastoma stem cells: driving resilience through Chaos. Trends Cancer 6, 223–235 10.1016/j.trecan.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B. et al. (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 6.Suvà M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., et al. (2014) Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahan P., Martinez Gala J., Delmas C., Monferran S., Malric L., Zentkowski D. et al. (2014) Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Cell Death Dis. 5, e1543 10.1038/cddis.2014.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broekman M.L., Maas S.L.N., Abels E.R., Mempel T.R., Krichevsky A.M. and Breakefield X.O. (2018) Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 14, 482–495 10.1038/s41582-018-0025-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asencio-Barría C., Defamie N., Sáez J.C., Mesnil M. and Godoy A.S. (2019) Direct intercellular communications and cancer: a snapshot of the biological roles of connexins in prostate cancer. Cancers (Basel) 11, 1370 10.3390/cancers11091370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osswald M., Jung E., Sahm F., Solecki G., Venkataramani V., Blaes J., et al. (2015) Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- 11.Weil S., Osswald M., Solecki G., Grosch J., Jung E., Lemke D. et al. (2017) Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol. 19, 1316–1326 10.1093/neuonc/nox070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung E., Osswald M., Blaes J., Wiestler B., Sahm F., Schmenger T., et al. (2017) Tweety-homolog 1 drives brain colonization of gliomas. J. Neurosci. 37, 6837–6850 10.1523/JNEUROSCI.3532-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto G., Brou C. and Zurzolo C. (2020) Tunneling nanotubes: the fuel of tumor progression? Trends Cancer 6, 874–888 10.1016/j.trecan.2020.04.012 [DOI] [PubMed] [Google Scholar]
- 14.Hekmatshoar Y., Nakhle J., Galloni M. and Vignais M.-L. (2018) The role of metabolism and tunneling nanotube-mediated intercellular mitochondria exchange in cancer drug resistance. Biochem. J. 475, 2305–2328 10.1042/BCJ20170712 [DOI] [PubMed] [Google Scholar]
- 15.Rustom A., Saffrich R., Markovic I., Walther P. and Gerdes H.-H. (2004) Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- 16.Sartori-Rupp A., Cordero Cervantes D., Pepe A., Gousset K., Delage E., Corroyer-Dulmont S. et al. (2019) Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 10, 342 10.1038/s41467-018-08178-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abounit S. and Zurzolo C. (2012) Wiring through tunneling nanotubes – from electrical signals to organelle transfer. J. Cell Sci. 125, 1089–1098 10.1242/jcs.083279 [DOI] [PubMed] [Google Scholar]
- 18.Abounit S., Bousset L., Loria F., Zhu S., de Chaumont F., Pieri L. et al. (2016) Tunneling nanotubes spread fibrillar α-synuclein by intercellular trafficking of lysosomes. EMBO J. 35, 2120–2138 10.15252/embj.201593411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S., Victoria G.S., Marzo L., Ghosh R. and Zurzolo C. (2015) Prion aggregates transfer through tunneling nanotubes in endocytic vesicles. Prion 9, 125–135 10.1080/19336896.2015.1025189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abounit S., Wu J.W., Duff K., Victoria G.S. and Zurzolo C. (2016) Tunneling nanotubes: a possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases. Prion 10, 344–351 10.1080/19336896.2016.1223003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Victoria G.S. and Zurzolo C. (2015) Trafficking and degradation pathways in pathogenic conversion of prions and prion-like proteins in neurodegenerative diseases. Virus Res. 207, 146–154 10.1016/j.virusres.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 22.Victoria G.S. and Zurzolo C. (2017) The spread of prion-like proteins by lysosomes and tunneling nanotubes: implications for neurodegenerative diseases. J. Cell Biol. 216, 2633–2644 10.1083/jcb.201701047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onfelt B., Nedvetzki S., Benninger R.K.P., Purbhoo M.A., Sowinski S., Hume A.N. et al. (2006) Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J. Immunol. 177, 8476–8483 10.4049/jimmunol.177.12.8476 [DOI] [PubMed] [Google Scholar]
- 24.Eugenin E.A., Gaskill P.J. and Berman J.W. (2009) Tunneling nanotubes (TNT) are induced by HIV- infection of macrophages. Cell Immunol. 254, 142–148 10.1016/j.cellimm.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souriant S., Balboa L., Dupont M., Pingris K., Kviatcovsky D., Cougoule C., et al. (2019) Tuberculosis exacerbates HIV-1 infection through IL-10/STAT3-dependent tunneling nanotube formation in macrophages. Cell Rep. 26, 3586–3599.e7 10.1016/j.celrep.2019.02.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korenkova O., Pepe A. and Zurzolo C. (2020) Fine intercellular connections in development: TNTs, cytonemes, or intercellular bridges? Cell Stress 4, 30–43 10.15698/cst2020.02.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignais M.-L., Caicedo A., Brondello J.-M. and Jorgensen C. (2017) Cell connections by tunneling nanotubes: effects of mitochondrial trafficking on target cell metabolism, homeostasis, and response to therapy. Stem Cells Int. 2017, 6917941 10.1155/2017/6917941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carone C., Genedani S., Leo G., Filaferro M., Fuxe K. and Agnati L.F. (2015) In vitro effects of cocaine on tunneling nanotube formation and extracellular vesicle release in glioblastoma cell cultures. J. Mol. Neurosci. 55, 42–50 10.1007/s12031-014-0365-9 [DOI] [PubMed] [Google Scholar]
- 29.Ding X., Ma M., Teng J., Teng R.K.F., Zhou S., Yin J. et al. (2015) Exposure to ALS-FTD-CSF generates TDP-43 aggregates in glioblastoma cells through exosomes and TNTs-like structure. Oncotarget 6, 24178–24191 10.18632/oncotarget.4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Formicola B., D'Aloia A., Dal Magro R., Stucchi S., Rigolio R., Ceriani M. et al. (2019) Differential exchange of multifunctional liposomes between glioblastoma cells and healthy astrocytes via tunneling nanotubes. Front. Bioeng. Biotechnol. 7, 403 10.3389/fbioe.2019.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L. and Zhang Y. (2015) Tunneling nanotubes between rat primary astrocytes and C6 glioma cells alter proliferation potential of glioma cells. Neurosci. Bull. 31, 371–378 10.1007/s12264-014-1522-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdebenito S., Audia A., Bhat K.P.L., Okafo G. and Eugenin E.A. (2020) Tunneling nanotubes mediate adaptation of glioblastoma cells to temozolomide and ionizing radiation treatment. iScience 23, 101450 10.1016/j.isci.2020.101450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avril T., Vauleon E., Hamlat A., Saikali S., Etcheverry A., Delmas C. et al. (2012) Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 22, 159–174 10.1111/j.1750-3639.2011.00515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubert C.G., Rivera M., Spangler L.C., Wu Q., Mack S.C., Prager B.C. et al. (2016) A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 76, 2465–2477 10.1158/0008-5472.CAN-15-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abounit S., Delage E. and Zurzolo C. (2015) Identification and characterization of tunneling nanotubes for intercellular trafficking. Curr. Protoc. Cell Biol. 67, 12.10.1–12.10.21 10.1002/0471143030.cb1210s67 [DOI] [PubMed] [Google Scholar]
- 36.Ferrick D.A., Neilson A. and Beeson C. (2008) Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today 13, 268–274 10.1016/j.drudis.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 37.Bates D., Mächler M., Bolker B. and Walker S. (2015) Fitting linear mixed-Effects models using lme4. J. Stat. Softw. 67, 1–48 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 38.Kuznetsova A., Brockhoff P.B. and Christensen R.H.B. (2017) Lmertest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 10.18637/jss.v082.i13 [DOI] [Google Scholar]
- 39.Deviers A., Ken S., Filleron T., Rowland B., Laruelo A., Catalaa I., et al. (2014) Evaluation of the lactate-to-N-acetyl-aspartate ratio defined with magnetic resonance spectroscopic imaging before radiation therapy as a new predictive marker of the site of relapse in patients with glioblastoma multiforme. Int. J. Radiat. Oncol. Biol. Phys. 90, 385–393 10.1016/j.ijrobp.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 40.Guo J., Yao C., Chen H., Zhuang D., Tang W., Ren G. et al. (2012) 681The relationship between Cho/NAA and glioma metabolism: implementation for margin delineation of cerebral gliomas. Acta Neurochir. (Wien) 154, 1361–1370 10.1007/s00701-012-1418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., et al. (2019) An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T. et al. (2004) Identification of human brain tumour initiating cells. Nature 432, 396–401 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- 43.Son M.J., Woolard K., Nam D.-H., Lee J. and Fine H.A. (2009) SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 4, 440–452 10.1016/j.stem.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhaak R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., et al. (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- 46.Zhu S., Bhat S., Syan S., Kuchitsu Y., Fukuda M. and Zurzolo C. (2018) Rab11a–Rab8a cascade regulates the formation of tunneling nanotubes through vesicle recycling. J. Cell Sci. 131, jcs215889 10.1242/jcs.215889 [DOI] [PubMed] [Google Scholar]
- 47.Osswald M., Jung E., Wick W. and Winkler F. (2019) Tunneling nanotube-like structures in brain tumors. Cancer Rep. 2, e1181 10.1002/cnr2.1181 [DOI] [Google Scholar]
- 48.Civita P., Leite M. and Pilkington G. J. (2019) Pre-clinical drug testing in 2D and 3D human in vitro models of glioblastoma incorporating non-neoplastic astrocytes: tunneling nano tubules and mitochondrial transfer modulates cell behavior and therapeutic response. Int. J. Mol. Sci. 20, 6017 10.3390/ijms20236017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linkous A., Balamatsias D., Snuderl M., Edwards L., Miyaguchi K., Milner T., et al. (2019) Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 26, 3203–3211.e5 10.1016/j.celrep.2019.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung E., Alfonso J., Osswald M., Monyer H., Wick W. and Winkler F. (2019) Emerging intersections between neuroscience and glioma biology. Nat. Neurosci. 22, 1951–1960 10.1038/s41593-019-0540-y [DOI] [PubMed] [Google Scholar]
- 51.Venkataramani V., Tanev D.I., Strahle C., Studier-Fischer A., Fankhauser L., Kessler T., et al. (2019) Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538 10.1038/s41586-019-1564-x [DOI] [PubMed] [Google Scholar]
- 52.Venkatesh H.S., Morishita W., Geraghty A.C., Silverbush D., Gillespie S.M., Arzt M., et al. (2019) Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 10.1038/s41586-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osswald M., Solecki G., Wick W. and Winkler F. (2016) A malignant cellular network in gliomas: potential clinical implications. Neuro Oncol. 18, 479–485 10.1093/neuonc/now014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Yu X., Xie C., Tan Z., Tian Q., Zhu D. et al. (2016) Rescue of brain function using tunneling nanotubes between neural stem cells and brain microvascular endothelial cells. Mol. Neurobiol. 53, 2480–2488 10.1007/s12035-015-9225-z [DOI] [PubMed] [Google Scholar]
- 55.Kretschmer A., Zhang F., Somasekharan S.P., Tse C., Leachman L., Gleave A., et al. (2019) Stress-induced tunneling nanotubes support treatment adaptation in prostate cancer. Sci. Rep. 9, 7826 10.1038/s41598-019-44346-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desir S., Wong P., Turbyville T., Chen D., Shetty M., Clark C., et al. (2019) Intercellular transfer of oncogenic KRAS via tunneling nanotubes introduces intracellular mutational heterogeneity in colon cancer cells. Cancers (Basel) 11, 892 10.3390/cancers11070892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connor Y., Tekleab S., Nandakumar S., Walls C., Tekleab Y., Husain A., et al. (2015) Physical nanoscale conduit-mediated communication between tumour cells and the endothelium modulates endothelial phenotype. Nat. Commun. 6, 8671 10.1038/ncomms9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laprie A., Catalaa I., Cassol E., McKnight T.R., Berchery D., Marre D. et al. (2008) Proton magnetic resonance spectroscopic imaging in newly diagnosed glioblastoma: predictive value for the site of postradiotherapy relapse in a prospective longitudinal study. Int. J. Radiat. Oncol. Biol. Phys. 70, 773–781 10.1016/j.ijrobp.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 59.Badr C.E., Silver D.J., Siebzehnrubl F.A. and Deleyrolle L.P. (2020) Metabolic heterogeneity and adaptability in brain tumors. Cell. Mol. Life Sci. 77, 5101–5119 10.1007/s00018-020-03569-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.