Abstract
It is the first report on complete chloroplast genome of Mikania micrantha (Asteraceae), a noxious invasive weed to South China. The genome is a circular molecule of 152,092 bp in length with 37.58% average GC content, and includes a large single copy region (83,793 bp), a small single copy region (18,287 bp), and two inverted repeat regions (25,006 bp). The M. micrantha cp genome encodes 135 unique genes, including 90 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. ML tree based on 16 complete cpDNA from Asteraceae indicated that M. micrantha has a close sister relationship with Ageratina adenophora and Praxelis clematidea. The complete cpDNA of M. micrantha provides useful molecular data for further phylogenetic and evolutionary analysis.
Keywords: Asteraceae, chloroplast genome, Mikania micrantha, noxious invasive weed
Mikania micrantha (Asteraceae) is one of the top 10 worst weeds in the world known as a mile-a-minute weed. It is an extremely fast growing, long-lived herbaceous vine. It was introduced to Hong Kong in 1884 (Zhang et al. 2004). After naturalization, M. micrantha has rapidly spread through the Pearl River Delta since 1984, due to lack of natural enemy (Huang et al. 2015). The species is a noxious invasive weed to South China. It not only kills other plants by blocking the light, twinning, smothering and preventing forest tree regeneration (Zhang et al. 2004), but also competes for water and nutrients with coexisting natives (Wang et al. 2004). Currently, studies on M. micrantha focus in investigation of basic features such as the biological characteristics (Hu & But 1994), photosynthetic activity (Wen et al. 2000), allelopathy (Shao et al. 2003), and genetic variation (Wang et al. 2008). However, its characteristics of chloroplast genome remain unresolved. Here, we reported the complete cp genome sequences of M. micrantha, which will contribute to elucidate adaptive evolutionary mechanism.
The leaves were from bamboo garden (N: 23°5'37.41”, E: 113°17'43.94”), Sun Yat-sen University, Guangdong Province, China, and the specimen is stored in Herbarium of Sun Yat-sen University (SYSU; voucher: L Huang 201408). We sequenced the complete cp genome by using Illumina HiSeqTM 2500 platform (Illumina Inc., San Diego, CA), which generated a total of 4.73 GB of 150-bp paired-end clean reads. Velvet (V 1.2.07) was used to assemble de novo sequences from clean reads (Zerbino & Birney 2008). A total length of 127,801 bp in ten chloroplast contigs were obtained through BLAST to the published invasive species cpDNA sequences in Asteraceae. PCR amplification was applied to bridge the gaps. The initial annotation of the assembled chloroplast genome was executed with Dual Organellar GenoMe Annotator (DOGMA) to predict protein coding genes, transfer RNA (tRNA) genes, and ribosome RNA (rRNA) genes. Furthermore, we performed BLASTN to determine accurate annotation of gene positions. Based on complete chloroplast sequences of 15 species corresponding to 15 different genera in Asteraceae, a maximum likelihood tree was constructed to explore phylogenetic relationship of M. micrantha using RAxMLGUI v.1.3 with 1000 bootstrap replicates (Silvestro & Michalak 2012). Trachelium caeruleum (Campanulaceae) was used as outgroup.
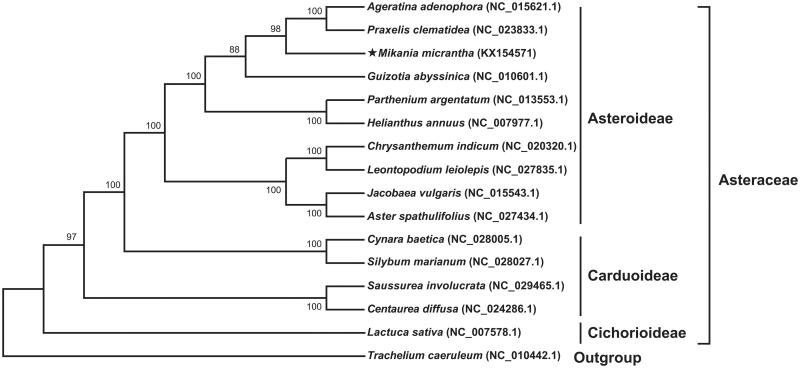
The complete cp genome of M. micrantha is a circular molecule of 152,092 bp in length with 37.58% average GC content (GenBank accession no. KX154571), and includes a large single copy region (LSC, 83,793 bp), a small single copy region (SSC, 18,287 bp), and two inverted repeat regions (IR, 25,006 bp). The M. micrantha cp genome contains 135 unique genes, including 90 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these genes, two (clpP and ycf3) contained two introns, which were located in LSC. In addition, 19 genes embraced one introns. ML result with 98% bootstrap showed that M. micrantha has a close sister relationship with Ageratina adenophora and Praxelis clematidea (Figure 1). The complete cpDNA of M. micrantha provides useful molecular data for further phylogenetic and evolutionary analysis.
Figure 1.
The ML phylogenetic tree based on complete chloroplast genome sequences of 15 species in Asteraceae and Trachelium caeruleum as outgroup. The complete chloroplast genome sequences were downloaded from GenBank, and the accession numbers were shown in the tree. ML bootstrap support values were indicated at the nodes.
Acknowledgments
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the National Natural Science Foundation of China [31370364, 31570652, and J1310025], the National Natural Science Foundation of Guangdong Province [2016A030313320], the Open Project of the State Key Laboratory of Biocontrol, China [SKLBC13KF07], Project of Department of Science and Technology of Shenzhen City, China [JCYJ20160425165447211].
References
- Hu YJ, But PPH.. 1994. A study on life cycle and response to herbicides of Mikania micrantha. Acta Sci Nat Univ Sunyatseni. 33:88–95. (in Chinese). [Google Scholar]
- Huang QQ, Shen YD, Li XX, Zhang GL, Huang DD, Fan ZW.. 2015. Regeneration capacity of the small clonal fragments of the invasive Mikania micrantha H.B.K: effects of the stolon thickness, internode length and presence of leaves. Weed Biol Manag. 15:70–77. [Google Scholar]
- Shao H, Peng SL, Zhang C, Xiang YC, Nan P.. 2003. Allelopathic potential of Mikania micrantha. Chin J Ecol. 22:62–65. (in Chinese). [Google Scholar]
- Silvestro D, Michalak I.. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337. [Google Scholar]
- Wang T, Su YJ, Chen GP.. 2008. Population genetic variation and structure of the invasive weed Mikania micrantha in southern China: consequences of rapid range expansion. J Hered. 99:22–33. [DOI] [PubMed] [Google Scholar]
- Wang BS, Wang YJ, Liao WB, Zan QJ, Li MG, Peng SL, Han AC, Zhang WY, Chen DP.. 2004. The invasion ecology and management of alien weed Mikania micrantha H.B.K. Beijing: Science Press; pp. 152–177. [Google Scholar]
- Wen DZ, Ye WH, Feng HL, Cai CX.. 2000. Comparison of basic photosynthetic characteristics between exotic invader weed Mikania micrantha and its companion species. J Tropi Substropi Bot. 8:139–146. (in Chinese). [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Ye WH, Cao HL, Feng HL.. 2004. Mikania micrantha H.B.K. in China-an overview. Weed Res. 44:42–49. [Google Scholar]