Abstract
Euphorbia kansui T.N. Liou ex S.B. Ho (Euphorbiaceae) is a perennial herb plant endemic to China. This species has important economic and medicinal values. In this study, we first characterized the complete nucleotide sequence of chloroplast (cp) genome of E. kansui using the Illumina Hiseq platform. The cp genome was 161,061 bp in length, comprising of a large single copy (LSC) region of 91,288 bp, a small single copy (SSC) region of 17,086 bp, and two inverted repeat regions of 26,343 bp each. The cp genome contains 130 genes, including 86 protein-coding genes, 8 ribosomal RNAs (rRNAs), and 36 transfer RNAs (tRNAs). The phylogenetic analysis indicated that E. kansui was placed as a sister to the congeneric Euphorbia esula.
Keywords: Chloroplast genome, conservation, Euphorbia kansui, phylogenetic analysis
Euphorbia kansui T.N. Liou ex S.B. Ho (Euphorbiaceae) is a perennial herb plant endemic to Northern China, and have long been used as traditional Chinese medicinal materials (Wang et al. 2003; Yan et al. 2014). In recent years, overexploitation by human beings, as well as the rapid climate change, results in the increasingly declining and fragmenting of its natural populations. It is thus urgent to take effective strategies to conserve and mange this endangered herb species. In plant, chloroplast (cp) DNA provided valuable phylogenetic information, owning to its conserved genome structures and comparatively high substitution rates (Wu and Ge 2012). Herein, we first characterize the complete chloroplast genome of E. kansui based on the Illumina next-generation sequencing technology.
The fresh leave tissues from a single individual of E. kansui were collected from Hechuan Town (Shaanxi, China; 110.3389°E, 35.1353°N), and genomic DNA was isolated using a modified CTAB method (Doyle and Doyle 1987). DNA sample and voucher specimen (No. EKLZH2013516) of E. kansui were deposited in the Northwest University Museum (NUM). Then, the high quality DNAs were subjected to Illumina sample preparation, and pair-read sequenced were conducted on the Illumina Hiseq 2500 platform (Novogene Bioinformatics Technology Co., Ltd). In total, about 1,260,648 high-quality reads were obtained and used for the complete cp genome reference-guided assembly using the MIRA 4.0.2 (Chevreux et al. 2004) and MITObim version 1.7 (Hahn et al. 2013). The annotation of cp genome was conducted using the online program Dual Organellar Genome Annotator (DOGMA, Wyman et al. 2004), and then manually adjusted the positions of start codes, stop codes, introns, and exons by comparison with homologous genes in other chloroplast genomes. In addition, all tRNA genes were further verified online using the software tRNAscan-SE1.21 (Schattner et al. 2005). Eventually, the annotated cp genome sequence of E. kansui has been submitted to GenBank with the accession number MH392274, and circular genome maps were drawn using the program OGDRAW (Lohse et al. 2013).
The complete cp genome of E. kansui was a typical quadripartite circular molecule with a length of 161, 061 bp, which comprising a large single copy (LSC) region of 91, 288 bp and a small single copy (SSC) region of 17, 086 bp, separated by two inverted repeat regions (IRs) of 26,343 bp. The plastid genome contains 130 genes, including 86 protein-coding genes, 36 transfer RNAs (tRNAs), and 8 ribosomal RNAs (rRNAs). Among them, 19 genes duplicated in the IRs. A total of 13 genes (atpF, rpoC1, rpl2, ndhB, ndhA, tRNA-Lys, tRNA-Leu, tRNA-Val, tRNA-Ala, and tRNA-Ile) contained one intron, and two genes (ycf3, rps12) contained two introns. The overall GC content of E. kansui plastome is 35.5%, while the corresponding values of LSC, SSC, and IR regions are 32.6%, 30.2%, and 42.4%, respectively.
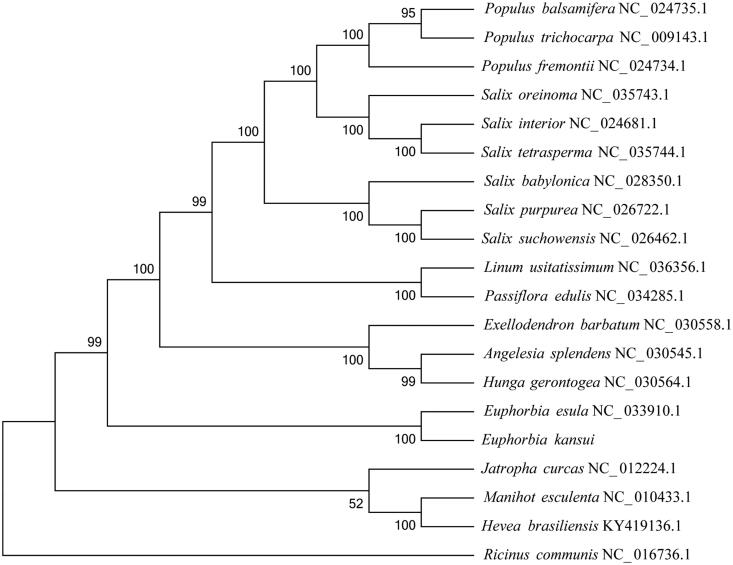
To investigate the phylogenetic position of E. kansui, the cp genomes of 19 species within order Malpighiales were downloaded from NCBI. All of the 20 complete plastome sequences were aligned using the software MAFFT (Katoh and Standley 2013) with the default parameters. The phylogenetic analysis was conducted using the program MEGA6 (Tamura et al. 2013) with 1000 bootstrap replicates. The results indicated that E. kansui was placed as a sister to the congeneric Euphorbia esula with high bootstrap value (Figure 1).
Figure 1.
Phylogenetic tree based on twenty complete chloroplast genome sequences.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Boil Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LY, Wang NL, Yao XS, Miyata S, Kitanaka S. 2003. Euphane and tirucallane triterpenes from the roots of Euphorbia kansui and their in vitro effects on the cell division of Xenopus. J Nat Prod. 66:630–633. [DOI] [PubMed] [Google Scholar]
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62:573–578. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yan XH, Fang MF, Qian ZQ, Liu ZL, Wang ML, Tian CP, Du MH, Li YJ, Li ZH, Zhao GF. 2014. Isolation and characterization of polymorphic microsatellites in the perennial herb Euphorbia kansui using paired-end Illumina shotgun sequencing. Conservation Genet Resour. 6:841–843. [Google Scholar]