Abstract
Phlaeoba infumata belongs to Acrididae, Orthoptera. The complete mitochondrial genome of P. infumata is 15,642 bp in length, and its arrangement was identical to the Locusta migratoria mitogenome, and contained 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and a A + T-rich region. Using the 13 PCGs and two rRNA of P. infumata, together with 10 other close-related and two outgroup species, we constructed Bayesian inference (BI) phylogenetic tree to validate the mitogenome sequences of P. infumata.
Keywords: Mitogenome, Phlaeoba infumata, phylogeny
Phlaeoba infumata belongs to Acrididae, Orthoptera. Phlaeoba which is a large genus of Acrididae distribute all over the world. 12 species of the genus are known at present. Among them, eight species are found in China, and distribute mainly in the south of China (Wei & Zheng 2005). The specimen of P. infumata was collected from Xing’an County (Guangxi, China) in 2006, and was now deposited in Molecular and Evolutionary Lab in Shaanxi Normal University in China. The PCR amplifications were performed following the study of Liu et al. (2006). The Staden Package 1.7 was used for the mitogenome assembly and annotation (Staden et al. 2000). The 17 tRNA genes were predicted using online software tRNAScan-SE 1.21 (Lowe & Eddy 1997), the remaining five tRNAs, two rRNA, 13 protein-coding genes (PCGs) and a A + T-rich region were identified and annotated by sequence alignment with Locusta migratoria mitogenome sequences (GenBank accession no. X80245.1).
The complete mitogenome of P. infumata is 15,642 bp in length and has been deposited in GenBank (accession no. KU866166). It consists of 13 PCGs, 22 tRNA genes, two rRNA genes and one A + T-rich region, and its arrangement was identical to the L. migratoria mitogenome. The overall base composition of the whole mitochondrial genome was A (42.61%), T (31.32%), G (10.84%) and C (15.23%) with an AT bias of 73.93%. The A + T-rich region is as long as 744 bp and contains the highest A + T content (83.07%). The length of tRNA genes ranged from 65 bp to 71 bp. The start codons of all PCGs were typical ATN except COI which started with CTT, and the stop codons of all the 13 PCGs were complete (TAA or TAG) except ND5 which ended with an incomplete stop condon (TA). The rrnL and rrnS genes are 1315 bp and 824 bp in length, respectively.
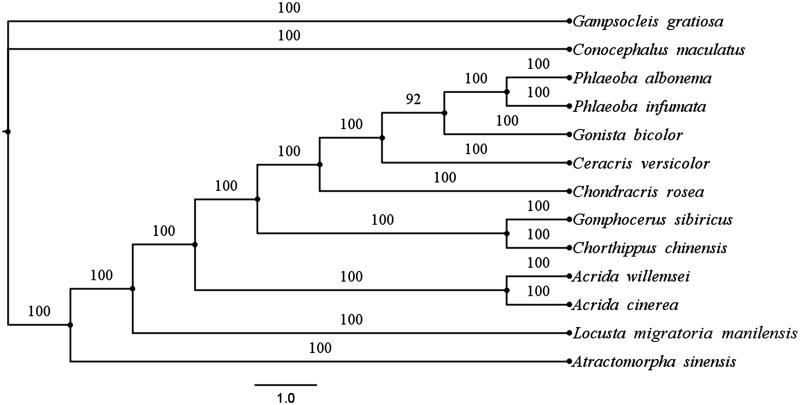
To furthermore validate the mitogenome of P. infumata, the phylogenetic analysis was peformed using MrBayes 3.2.2 based on concatenated mitogenome dataset (PCGs and rRNAs) of P. infumata in this study (Ronquist & Huelsenbeck 2003), and together with other 12 species from GenBank, including 10 close-related species and two ensiferan outgroup species (Figure 1).
Figure 1.
The BI phylogenetic tree of Phlaeoba infumata and other 12 species including 10 close-related species and two outgroup species based on mitochondrial PCGs and rRNAs concatenated dataset. GenBank accession nos.: Phlaeoba albonema NC_011827.1; Atractomorpha sinensis NC_011824.1; Locusta migratoria manilensis NC_014891.1; Chondracris rosea NC_019993.1; Acrida willemsei NC_011303.1; Gomphocerus sibiricus NC_021103.1; Chorthippus chinensis NC_011095.1; Acrida cinerea NC_014887.1; Conocephalus maculatus NC_016696.1; Ceracris versicolor KJ188251.1; Gampsocleis gratiosa EU527333.1; Gonista bicolor NC_029205.1
Disclosure statement
The authors report no conflicts of interest.
Funding information
This work was supported by the China Postdoctoral Science Foundation under Grant no. 2014M562369; Shaanxi Xueqian Normal University Scientific Research Foundation for the introduction of talent under Grant no. 2014DS008; and Shaanxi Xueqian Normal University Scientific Research Foundation under Grant no. 2015YBKJ024.
References
- Liu N, Hu J, Huang Y.. 2006. Amplification of grasshoppers complete mitochondrial genomes using long PCR. Chin J Zool. 41:61–65 (in Chinese with an English abstract). [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence . Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK.. 2000. The Staden package, 1998. Methods Mol Biol. 132:115–130 . [DOI] [PubMed] [Google Scholar]
- Wei S, Zheng Z.. 2005. A study on the C-banding karyotypes of two species of oedaleus fieber in Guangxi (Acridoidea: Acrididae). J South China Norm Univ (Nat Sci Ed). 2:18–22 (in Chinese with an English abstract). [Google Scholar]