Abstract
We determined the complete chloroplast genome sequences of Aconitum austrokoreense Koidz., an endangered endemic species in Korea. The chloroplast DNA is 155,682 bp in length and encodes 37 tRNAs, 8 rRNAs, and 86 protein-coding genes. Phylogenetic analysis and sequence comparison of protein-coding genes with those in other Ranunculaceae chloroplast DNAs showed that the chloroplast genome of A. austrokoreense is closely related to that of A. chiisanense and large sequence variations identified in rps16, matK, and rpl20 are specific to these two species.
Keywords: Aconitum austrokoreense, endemic, chloroplast genome, sequence variation
Aconitum austrokoreense Koidz. is a member of genus Aconitum in Ranunculaceae and can be easily distinguished from other species in the genus by undivided or indistinctly 3-lobed leaves, yellowish white flowers with light violet margin, and pubescent pedicels and ovaries (Kadota 1987). Aconitum austrokoreense is an endemic species in Korea showing a very limited distribution with a few populations in rocky habitats near the trails along the mountain valleys (Kadota 1987; Park 2007). In Eastern Asia, the Aconitum species have been well known for their toxicity and medicinal effect (Lee 1979; Tang et al. 2012). In particular, the dried tuber of A. austrokoreense along with other Aconitum species has been commonly used as traditional herbal medicine to treat symptoms such as atonia, inflammation, paralysis, and coldness of the extremities (Lee et al. 1997). In recent years, natural A. austrokoreense populations continue to decline due to its limited distribution, over-harvesting for medicinal use, and habitat loss (Suh et al. 2004). For this reason, it is currently classified as a vulnerable species on the Red Data Book of Endangered Vascular Plants in Korea (Suh & Kim 2012) and has been protected as Level II Endangered Wild Plants by the Ministry of Environment, Korea.
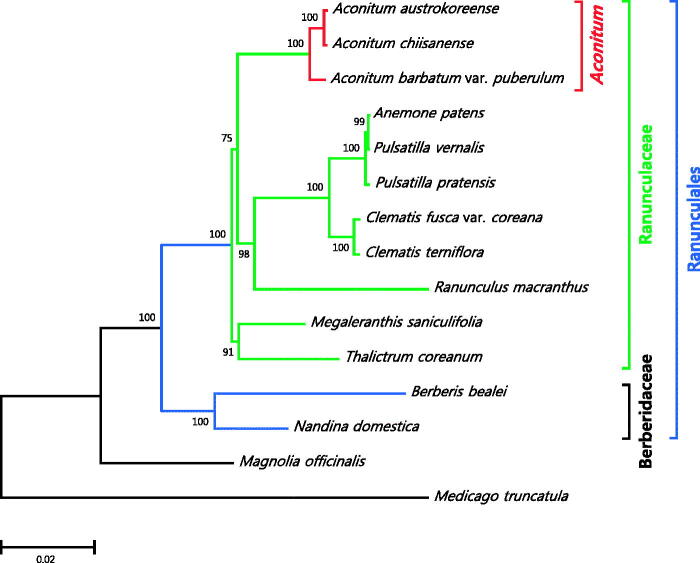
In this study, we determined the complete chloroplast (cp) DNA sequence of A. austrokoreense that will be a valuable resource for genetic and phylogenetic study of this species. The specimen was collected from the valleys of Mt. Chiri and deposited in National Institute of Biological Resources with the accession number NIBR-VP0000494177. Sequencing was performed using the Illumina MiSeq platform (Illumina Inc., San Diego, CA) and a total of 28 million paired-end reads were generated and assembled into a circular DNA (Genbank accession KT820663). The cpDNA was 155,682 bp in length and consisted of two IRs (25,613 bp), one LSC (86,910 bp), and one SSC (17,546 bp). Annotation using DOGMA (Wyman et al. 2004) and Geneious (R7.1.8; Kearse et al. 2012) predicted a total of 131 genes (37 tRNA, 8 rRNA, and 86 protein-coding genes), of which 18 genes (7 tRNA, 4 rRNA, and 7 protein-coding genes) were duplicated into two IRs. A total of 22 genes (8 tRNA and 14 protein-coding genes) contained one or two introns. The maximum likelihood tree constructed using 79 non-redundant protein-coding genes in cpDNAs from 12 Ranunculaceae species and two outgroup species indicated that the cp genome of A. austrokoreense is closely related to the cp DNA of A. chiisanense that we reported recently (Lim et al. 2015; Figure 1). Furthermore, sequence comparison identified several distinct sequence variations in protein-coding genes of Aconitum species. A large deletion within ycf2 and loss of rpl32 were evident in all Aconitum species, whereas truncation of rps16 and matK and insertion in rpl20 were specific to A. austrokoreense and A. chiisanense. The unique sequence divergence identified in the cpDNA of A. austrokoreense will serve as a molecular basis for conservation and maintenance of this species as well as phylogenetic study of Aconitum and Ranunculaceae.
Figure 1.
Maximum likelihood tree based on the chloroplast protein-coding genes of 15 taxa including Aconitum austrokoreense. Nucleotide sequences of 75 non-redundant conserved protein-coding genes from order Ranunculales as well as distantly related taxa were aligned and used for maximum likelihood phylogenetic analysis in MEGA7 with bootstrap test of 1000 replications. The NCBI accession numbers of chloroplast DNA sequences used in this study are as follows: Aconitum austrokoreense (KT820663), A. barbatum var. puberulum (KC844054), A. chiisanense (KT820665), Anemone patens (KR297057), Berberis bealei (NC022457), Clematis fusca var. coreana (KM652489), C. terniflora (NC028000), Megaleranthis saniculifolia (NC012615), Nandina domestica (NC008336), Pulsatilla pratensis (KR297059), P. vernalis (KR297061), Ranunculus macranthus (DQ359689), Thalictrum coreanum (KM206568), Magnolia officinalis (NC020316), and Medicago truncatula (NC003119). The scale bar represents the number of substitutions per site.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Kadota Y. 1987. A revision of Aconitum subgenus Aconitum (Ranunculaceae) of East Asia. Sanwa Shoyaku, Utsunomiya, Tokyo, Japan. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TB. 1979. Illustrated flora of Korea. Seoul, Korea: Hyangmunsa. [Google Scholar]
- Lee MT, Sung HK, Whang WK, Kim IH.. 1997. Alkaloids from the tuber of Aconitum chiisanense. Yakhak Hoeji. 31:161–173. [Google Scholar]
- Lim CE, Kim GB, Baek S, Han S-M, Yu H-J, Mun J-H.. 2015. The complete chloroplast genome of Aconitum chiisanense Nakai (Ranunculaceae). Mitochondrial DNA. Doi: 10.3109/19401736.2015.1110805. [DOI] [PubMed] [Google Scholar]
- Park C-W. 2007. Aconitum In: Flora of Korea Editorial Committee, editor. The genera of vascular plants of Korea. Seoul, Korea: Academy Publishing Co. [Google Scholar]
- Suh Y, Kim YJ.. 2012. Korean red list of threatened species: mammals, birds, reptiles, amphibians, fishes and vascular plants. Incheon, Korea: National Institute of Biological Resources. [Google Scholar]
- Suh MH, Koh KS, Ku YB, Kil JH, Choi TB, Suh SU, Oh HK, Lee IK, Na JG, Hyun JO, Ph JG. 2004. The conservation strategy for the endangered and reserved plants based on the ecological and genetic characteristics (IV). Incheon, Korea: National Institute of Environmental Research. [Google Scholar]
- Tang Q-F, Ye W-C, Liu J-H, Yang C-H.. 2012. Three new hetisine-type diterpenoid alkaloids from Aconitum coreanum. Phytochem Lett. 5:397–400. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]