Abstract
The complete mitochondrial genome of the endangered Chinese black-necked pond turtle, Mauremys nigricans, was determined in this study. The mitogenome was 16 779 bp in length, and the GC content was 39.07%. The genomes encoded 37 genes typically found in other vertebrates. In the control region, three CSBs were identified, followed by long tandem repeats of (ATATCATATT)10(ATATC)(ATATATC)7(TA)102. The mitochondiral genome sequence would be useful for the phylogenetic and conservation studies of Asian endangered turtles.
Keywords: Chinese black-necked pond turtle, Mauremys nigricans, mitochondrial genome
The Chinese black-necked pond turtle, Mauremys nigricans, is native to the southern region of China, and listed in Appendix III of CITES and as endangered in the IUCN red list of threatened species (Asian Turtle Trade Working Group 2000). It is close to M. reevesii in appearance. But the two lateral keels are not obvious, and red color appears on the head, neck and limbs in male individuals, which are distinctive characters from M. reevesii (Shi 2008).
A wild living male turtle was caught by a local farmer and conserved in Guangdong province, China. The nails of the specimen were used to extract genomic DNA. A library was constructed and sequenced using Illumina Hiseq2500. Mitochondrial reads were captured from high quality data by blasting to the mitchondrial sequences of other Mauremys species. The complete mitchondrial genome was assembled and the arrangement and characterization of the genome were determined using the program DOGMA, UT Austin (Wyman et al. 2004; http://dogma.ccbb.utexas.edu/) and tRNA scan-SE 1.21, Santa Cruz, CA (Schattner et al. 2005; http://lowelab.ucsc.edu/tRNA Scan-SE).
The complete mitogenome is 16 779 bp in size, and the GC content is 39.07%. Thirty-seven genes are encoded in the genome, of which 13 genes are protein-coding, 2 genes are rRNA and 22 genes are tRNA. The control region is 1253 bp in length, containing three conserved sequence boxes (CSB1, CSB2 and CSB3), and a long tandem repeat (VNTR). The VNTR is 358 bp long, consisting of 10 repeats of (ATATCATATT), 1 (ATATC), 7 repeats of (ATATATC) and 102 repeats of (TA).
A maximum likelihood (ML) analyses (Figure 1) were conducted with congeneric species distributed in Asia using RAxML8.1.5 software, Lausanne, Switzerland (Stamatakis 2006). The reconstructed phylogeny indicated the M. nigricans was close to the M. reevesii and M. sinensis. The barcoding sequence, a 650-bp region of COI gene, which was used for species identification in many animals (Hebert et al. 2003; Reid et al. 2011), was analyzed using MEGA version 6, Philadelphia, PA (Tamura et al. 2013). The Kimura 2-parameter (K2P) distance of this specimen to the voucher specimen (MVZ 130463; Parham et al. 2001) is 0.002, and 0.025 to M. reevesii (FJ469674), with which the two close species could be distinguished.
Figure 1.
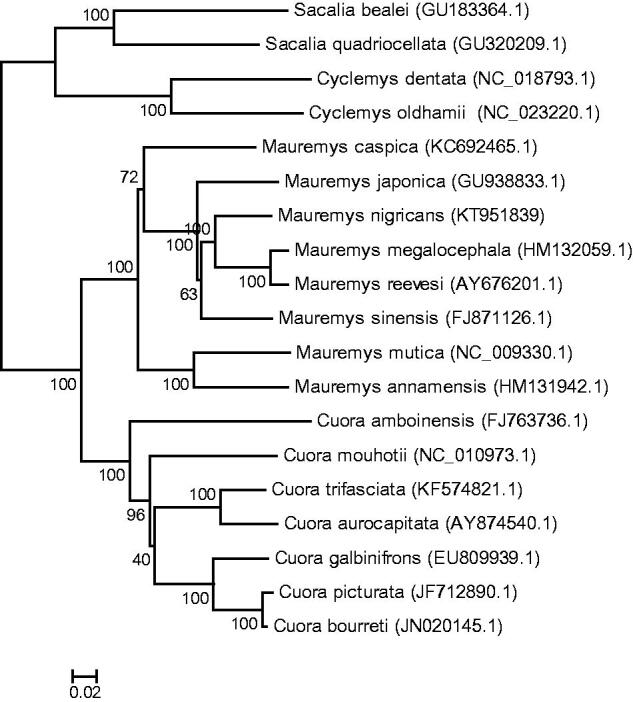
Phylogenetic analysis (ML topology) of M. nigricans based on the whole mitochondrial genome sequences. Numbers at each node represent the bootstrap value for ML analysis.
Nucleotide sequence accession number
The complete mitochondrial genome of M. nigricans has been assigned Genbank accession number KT951839.
Disclosure statement
This research was supported by the National Science and Technology Basic Research Program of China (2013FY110700).
References
- Asian Turtle Trade Working Group 2000. Mauremys nigricans. The IUCN Red List of Threatened Species :e.T4656A11059086. [Google Scholar]
- Hebert PD, Ratnasingham S, deWaard JR.. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 270:S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham JF, Simison WB, Kozak KH, Feldman CR, Shi HT. 2001. New Chinese turtles: endangered or invalid? A reassessment of two species using mitochondrial DNA, allozyme electrophoresis and known‐locality specimens[J]. Anim Conserv. 4:357–367. [Google Scholar]
- Reid BN, Le M, Mccord WP, Iverson JB, Georges A, Bergmann T, Amato G, Desalle R, Naro-Maciel E.. 2011. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol Ecol Resour. 11:956–967. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33 (suppl 2):W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HT. 2008. Identification manual for traded turtles in China. Beijing: Encyclopedia of China Publishing House. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]


