Abstract
Huperzia javanica (Sw.) C. Y. Yang is a valuable medical herb used for treating Alzheimer’s disease. Here, we described the complete chloroplast genome of H. javanica using Illumina paired-end sequencing. The total genome length is 154,415 bp, containing 119 unique genes, with 86 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. The gene content and their order are consistent with two previously reported Huperzia genomes. The overall GC content of the chloroplast genome of H. javanica is 36.4%. The topology of our maximum-likelihood tree is consistent with topologies found in previous studies, with H. javanica sister to a clade of H. serrata and H. lucidula. We support the recognition of H. javanica as an independent species. Huperzia serrata is more closely related to H. lucidula than to H. javanica.
Keywords: Huperzia javanica, Huperzia, Lycopodiaceae, chloroplast genome
Huperzia javanica C. Y. Yang (Lycopodiaceae) is a perennial, evergreen, South East Asian herb found in South China, India, Japan, South Korea, and the Philippines (Sun 2015). It grows on shady places in humid forests and valleys at an elevation ranging from 300–1200 m (Shrestha & Zhang 2015; Sun 2015). Huperzia species are known as valuable medicinal herbs. Especially, H. javanica and H. serrata are used for the treatment of Alzheimer’s disease (Tang 1996; Wang et al. 1998; Yang et al. 2008). Recently, H. javanica was distinguished as independent species from H. serrata complex through a previous study using morphological and climatic data (Shrestha & Zhang 2015), but we could not distinguish these species on the dried condition for making the oriental medicine based on only morphology. Chloroplast genomes are widely used in molecular phylogeny, to assess genetic diversity in conservation genetics, and in molecular identification such as DNA barcoding (Burke et al. 2012; Huang et al. 2014; Walker et al. 2014). In this study, we describe the chloroplast genome of H. javanica and reveal the phylogenetic relationship between H. javanica and H. serrata, thus contributing to the molecular identification and the conservation biology of H. javanica. The annotated sequence was deposited in GenBank under accession number KY609860.
Leaf material of H. javanica was collected in Shaoguan, Guangdong province in south China. The voucher specimen (R. Wei CBL011) has been deposited in the Herbarium of the Institute of Botany, Chinese Academy of Sciences (PE). Total DNA was extracted from silica gel dried leaves with a modified cetyltrimethylammonium bromide (CTAB) method (Li et al. 2013). The NEBNext DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) was used for library construction. Paired-end reads of 2 × 150 bp then were generated using an Illumina HiSeq PE150 (Illumina, San Diego, CA). A total of 8,688,705 paired-end sequence reads of 150 bp were generated, of which 230,793 paired-ends reads belong to the chloroplast genome. The chloroplast genome data were extracted using H. lucidula (AY660566) as a reference and assembled de novo with Geneious v. R9.0.5 (Kearse et al. 2012). The initial annotation of H. javanica chloroplast genome was performed using Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. 2004). After initial annotation, the putative starts, stops, and intron positions were determined by comparison with homologous genes in previously reported Huperzia chloroplast genomes. The tRNA genes were annotated using DOGMA and tRNAscan-SE (Schattner et al. 2005). The circular chloroplast genome map was drawn using the OGDraw program (Lohse et al. 2013).
The complete size of the H. javanica chloroplast genome is 154,415 bp, which includes a pair of inverted repeat (IR) regions of 15,315 bp separated by a small single copy (SSC) region of 19,667 bp and a large single copy (LSC) region of 104,120 bp similar to the previously reported Huperzia chloroplast genomes (Wolf et al. 2005; Guo et al. 2016). The H. javanica chloroplast genome contains 119 genes, 11 of which are duplicated in the IR region, giving a total of 130 genes. The chloroplast genome of H. javanica contains 29 distinct tRNAs, four of which are duplicated in the IR region. 13 genes (atpF, ndhA, ndhB, petB, petD, rps12, rps16, rpl2, rpl16, rpoC1, trnA-UGC, trnL-UAA, trnV-UAC) contain one intron, while two genes (clpP, ycf3) contain two introns. The rps12 gene is divided in two independent transcription units. The overall GC content is 36.4%, in particular 34.5% in the LSC, 32.9% in the IR, and 45.0% in the SSC region.
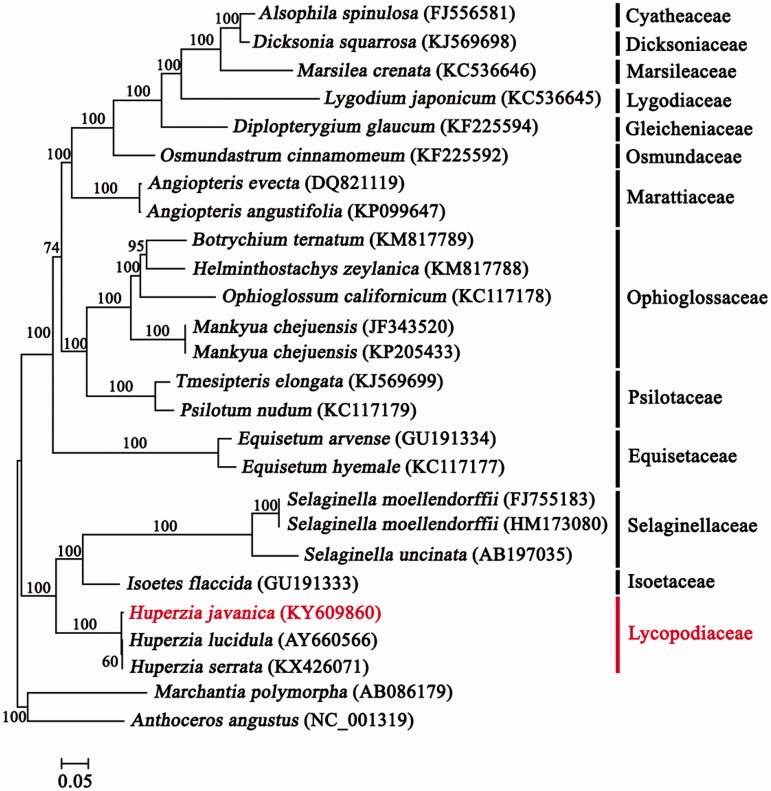
Phylogenetic analysis based on 72 protein-coding genes was performed using extra species from Lycopodiaceae to Cyatheales in the PPG I system (PPG I 2016), and two Bryophyte species (Anthoceros angustus and Marchantia polymorpha) as the outgroup (Figure 1). A total of 55,034 bp was aligned using MAFFT (Katoh et al. 2002). Maximum-likelihood (ML) analysis was performed using RAxML v. 7.4.2 with 1000 bootstrap replicates and the GTR + I + G model (Stamatakis 2006; Darriba et al. 2012). Our ML tree topology is consistent with topologies published in previous studies (Lu et al. 2015; PPG I 2016) (Figure 1). Although H. javanica is morphologically more similar to H. serrata, the phylogenetic relationship of H. serrata is closer to that of H. lucidula than to that of H. javanica (Figure 1). Moreover, this is confirmed by the sequence variation rates of the data matrix of 72 aligned protein-coding gene sequences of these three Huperzia species, which show that the sequence variation rate between H. serrata and H. javanica (0.411%) is slightly higher than that between H. serrata and H. lucidula (0.099%). These results are consistent with those of a previous study using morphological and climatic data, in which H. javanica was separated from H. serrata and treated as independent species (Shrestha & Zhang 2015).
Figure 1.
Phylogenetic tree based on 72 protein-coding genes using the ML method. Taxon in bold is the new genome reported in this study. Bootstrap values are shown above the nodes.
Acknowledgments
The authors thank Ran Wei at the Institute of Botany, Chinese Academy of Sciences for help with collecting the sample.
Disclosure statement
The authors report no conflicts of interest, and are solely responsible for the content and writing of this paper.
References
- Burke SV, Grennan CP, Duvall MR.. 2012. Plastome sequences of two New World bamboos-Arundinaria gigantea and Cryptochloa strictiflora (Poaceae)-extend phylogenomic understanding of Bambusoideae. Am J Bot. 99:1951–1961. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZY, Zhang HR, Shrestha N, Zhang XC.. 2016. Complete chloroplast genome of a valuable medicinal plant, Huperzia serrata (Lycopodiaceae), and comparison with its congener. Appl Plant Sci. 4:1600071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ.. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang S, Yu J, Wang L, Zhou S.. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78. [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JM, Zhang N, Du XY, Wen J, Li DZ.. 2015. Chloroplast phylogenomics resolves key relationships in ferns. J Syst Evol. 53:448–457. [Google Scholar]
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54:563–603. [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha N, Zhang XC.. 2015. Recircumscription of Huperzia serrata complex in China using morphological and climatic data. J Syst Evol. 53:88–103. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Sun BY. 2015. Lycopodiaceae In: Park CW, editors. Flora of Korea, vol. 1 Incheon, South Korea: National Institute of Biological Resources; p. 13–16. [Google Scholar]
- Tang XC. 1996. Huperzine A (shuangyiping): a promising drug for Alzheimer's disease. Zhongguo Yao Li Xue Bao. 17:481–484. [PubMed] [Google Scholar]
- Walker JF, Zanis MJ, Emery NC.. 2014. Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am J Bot. 101:722–729. [DOI] [PubMed] [Google Scholar]
- Wang L, Ji X, Wong Q, Yang J.. 1998. Observations of 36 cases on Alzheimer’s disease treatment using Huperzine A. J Nantong Med Coll. 18:486–488. [Google Scholar]
- Wolf PG, Karol KG, Mandoli DF, Kuehl J, Arumuganathan K, Ellis MW, Mishler BD, Kelch DG, Olmstead RG, Boore JL.. 2005. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene. 350:117–128. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Huang H, Chen DJ, Zhang JC.. 2008. Ultrasonic extraction and determination of Huperzine A from H. Javanica in Sanming region. Appl Chem Ind. 12:008. [Google Scholar]