Abstract
The complete mitochondrial genome of Rhynchocinetes durbanensis (Rhynchocinetidae: Rhynchocinetes) was sequenced in this study. The genome sequence was 17,695 bp in size, with the base composition of 35.14% A, 32.98% T, 20.34% G and 11.55% C of the light strand. The gene order and genes were the same as that found in other previously reported shrimps, including 13 protein-coding genes, 24 transfer RNA genes and two ribosomal RNA genes. Except for ND5, ND4, ND4L, ND1 genes and eight tRNA genes and two ribosomal RNA genes, all other mitochondrial genes were encoded on the heavy strand. The start codon of COX1 gene was not determined. These complete mitogenome data provide the basis for taxonomic and conservation research of Rhynchocinetes durbanensis, and closely related species.
Keywords: Complete mitochondrial genome, Rhynchocinetes durbanensis, shrimp
Rhynchocinetes durbanensis, found in the Indo-Pacific, is commonly known as the hingebeak prawn, which belongs to the family Rhynchocinetidae. It has many “Y”-shape white spots on the upper front part of its carapace and have features red and white lines on a translucent body (Calado 2009). Rhynchocinetes durbanensis was first described scientifically by Isabella Gordon in 1936 (Gordon 1936; De Grave, 2012). Rhynchocinetes durbanensis has been frequently confused with R. uritai (Okuno & Takeda 1992), and also closely resembles R. brucei (Ferrari & Ferrari 2002). As the development of marine aquarium, R. durbanensis is popular in the aquarium trade. In this study, we first determined the complete mitochondrial genome of R. durbanensis, which contribute to provide basic molecular data to the study on its systematics and conservation biology.
The R. durbanensis (RD150801) was collected from Hua Di Wang Flower & Aquarium Market in Guangzhou, Guangdong province in 2015 (23°7'12.00″N 113°15'0.00″E). The specimen stored in Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences. Genomic DNA of shrimp tissues was extracted by blood and cell culture DNA kit (QIAGEN, Hilden, FL) and then sequenced using Illumina’s HiSeq 2500 platform (Illumina Inc., San Diego, CA) with 500 bp insert-size DNA library and a pair-end 125 bp sequencing strategy (Miller et al. 2010).
The complete mitochondrial genome of R. durbanensis is 17,695 bp in length with an overall base composition of 5835 A (32.98%), 6218 T (35.14%), 2043 G (11.54%) and 3599 C (20.34%) (GenBank accession no. KT590405). The mitochondrial genome of R. durbanensis is a circular molecule containing 13 PCGs, 24 tRNA genes and two rRNA genes. 12S-rRNA and 16S-rRNA were located between the TrnaLeu and ATP8 genes and separated by the tRNAVal gene, at the length of 1362 bp and 824 bp, respectively. Most of the 39 genes are encoded on the heavy strand (H-strand) except ND5, ND4, ND4L, ND1 genes, eight tRNA genes and two ribosomal RNA genes, which encoded on the light strand. All tRNA genes can fold into a typical cloverleaf structure, with length ranges from 59 to 70 bp. Five of the 13 protein-coding genes (COX2, ATP6, ND4L, CYTB and ATP8) start with ATG, four of the 13 protein-coding genes (COX3, ND3, ND5 and ND2) start with ATT, ND6 and ND1 start with ATA, ND4 start with GTG, whereas the initiation codon of COX1 could not been determined. This phenomenon also been found in other shrimps, such as Rimicaris kairei (Yang et al. 2012), Upogebia pusilla (Shen et al. 2013) and Farfantepenaeus californiensis (Shen et al. 2007). All protein-coding genes harboured the typical termination codon TAA.
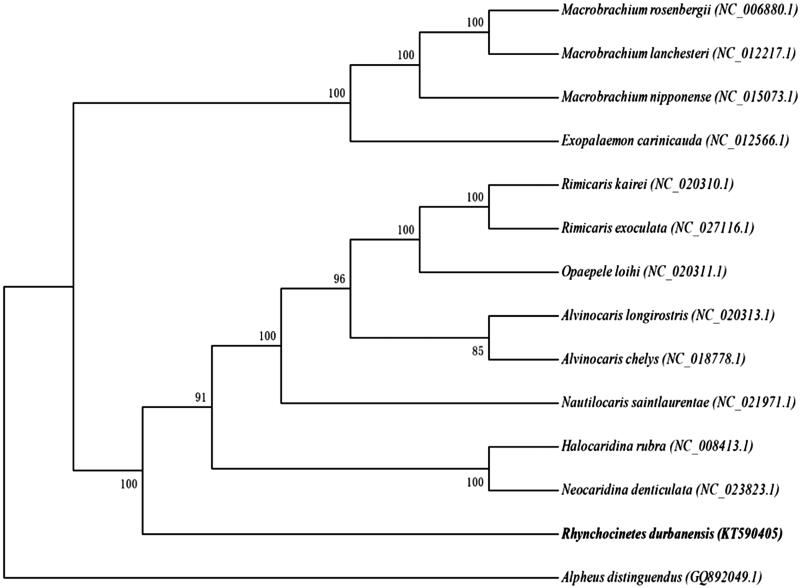
Some available mitochondrial genome sequences of the congeneric species were used in phylogenetic analysis carried out by maximum-likelihood (ML) method (Figure 1) (Abascal et al. 2005; Posada 2008; Tamura et al. 2011). As expected, R. durbanensis was joined with Neocaridina denticulata and Halocaridina rubra to form a tribe. The reconstructed phylogeny indicated that R. durbanensis was closely related to N. denticulata and H. rubra.
Figure 1.
Phylogenetic analysis (ML topology) of R. durbanensis based on the whole mitochondrial genome sequences. Numbers at each node represent the bootstrap value for ML analysis.
Acknowledgements
The authors thank Science Corporation of Gene for bioinformatics analysis support.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by grant from Henan Key Scientific and Technological Project (152102110091).
References
- Abascal F, Zardoya R, Posada D.. 2005. ProtTest: selection of best-fit models of protein evolution. C Bioinform. 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Calado R. 2009. Marine ornamental shrimp: biology, aquaculture and conservation. New York: John Wiley & Sons. [Google Scholar]
- De Grave S. 2012. Rhynchocinetes durbanensis Gordon, 1936. World Register of Marine Species. Retrieved 2012 Nov 17.
- Ferrari A, Ferrari A.. 2002. “Reef Life”. Firefly Books. New York: John Wiley & Sons; p. 264. [Google Scholar]
- Gordon I. 1936. On the macruran genus Rhynchocinetes, with description of a new species. Proc Zool Soc Lond. 106:75–88. [Google Scholar]
- Miller JR, Koren S, Sutton G.. 2010. Assembly algorithms for next generation sequencing data. Genomics. 95:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno J, Takeda M.. 1992. Distinction between two hinge-beak shrimps, Rhynchocinetes durbanensis Gordon and R. uritai Kubo (Family Rhynchocinetidae). Rev Francaise d'aquariologie herpetol. 19:85–90. [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Shen H, Braband A, Scholtz G.. 2013. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol Phylogenet Evol. 66:776–789. [DOI] [PubMed] [Google Scholar]
- Shen X, Ren J, Cui Z, Sha Z, Wang B, Xiang J, Liu B.. 2007. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene. 403:98–109. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lu B, Chen DF, Yu YQ, Yang F, Nagasawa H, Yang WJ.. 2012. When did decapods invade hydrothermal vents? Clues from the Western Pacific and Indian Oceans. Mol Biol Evol. 30:224. [DOI] [PubMed] [Google Scholar]