Abstract
Eggplant Solanum melongena L. is one of the most economically important vegetable crops. Here, we report the complete chloroplast (cp) genome of eggplant. The cp genome size was 154,289 bp that contained a pair of IR regions of 25,566 bp, one large single-copy (LSC) of 84,749 bp and a small single-copy (SSC) of 18,408bp, respectively. It encoded 125 predicted unique functional genes, including 84 tRNA genes, 85 protein-coding genes and 8 rRNA genes. The GC content was 37.86%. Phylogenetic analysis clearly showed a close evolutionary relationship between S. melongena and other species in the genus Solanum. The complete chloroplast genome of S. melongena provides valuable data for genetic improvement and allele mining of eggplant germplasm.
Keywords: Chloroplast genome, eggplant, phylogenomic analysis; Solanum melongena
Eggplant (Solanum melongena L.) is one of the most economically valuable vegetable species belongs to Solanaceae. It is an important member of the largest genera Solanum that consists of more than 1000 plant species. Compared with the other two Solanum model species, tomato and potato, eggplant possesses several agronomically unique traits including extra-large fruit size, high tolerance to biotic and abiotic stresses and parthenocarpy without any negative pleiotropic effects (Sakata et al. 1996; Saito et al. 2009). Fruits of eggplant are rich in nutrition contents such as vitamins that have recently been reconsidered as a good source of free radical scavengers, such as anthocyanins and phenolics (Stommel & Whitaker 2003; Azuma et al. 2008). The increase of genetic information in eggplant will not only enhances comparative studies of genetics, physiology, development, and evolution of solanaceous plants, but also accelerates novel allele mining in abundant eggplant germplasm (Daunay and Lester 1988).
Here, we assembled and characterized the complete chloroplast (cp) genome of S. melongena from the whole genome sequencing project of eggplant (Hirakawa et al. 2014). The sequenced S. melongena was a purebred cultivar (‘Nakate-Shinkuro’), one of the founders of modern commercial cultivars and a typical Asian cultivars as well. The cp genome readings of eggplant were filtered by Blastn (Altschul et al. 1990) using the S. nigrum cp genome sequence as a reference (Khan et al. 2015). About 813.5 Mb chloroplast readings were obtained and subjected to SOAPdenovo (Li et al. 2009) to assemble into the complete cp genome. Annotation was performed with Dual Organellar Geno Me Annotator (DOGMA, UT Austin, http://dogma.ccbb.utexas.edu). Ribosome RNA (rRNA), protein-coding and transfer RNA (tRNA) genes were predicted using default parameters (Wyman et al. 2004). The complete cp genome sequence with all annotated genes was deposited into GenBank under the accession number of KU682719. Thirteen complete cp genomes in Solanaceae were sampled for phylogenetic analysis. MAFFT (version 7.294, http://mafft.cbrc.jp/alignment/software/.) was used for whole-genome alignments. Maximum-likelihood (ML) analysis was performed by using ‘fast bootstrap’ algorithm under GTRGAMMAX model, replicated 100 times.
The complete cp genome of S. melongena exhibited a circular DNA molecule of 154,289 bp with a typical quadripartite structure. It consisted of a pair of IR regions of 25,566 bp, one large single-copy (LSC) of 84,749 bp and a small single-copy (SSC) of 18,408 bp. The cp genome encoded 125 unique functional genes, including 84 tRNA genes, 85 protein-coding genes and 8 rRNA genes. In addition, the GC content of the cp genome was 37.86%.
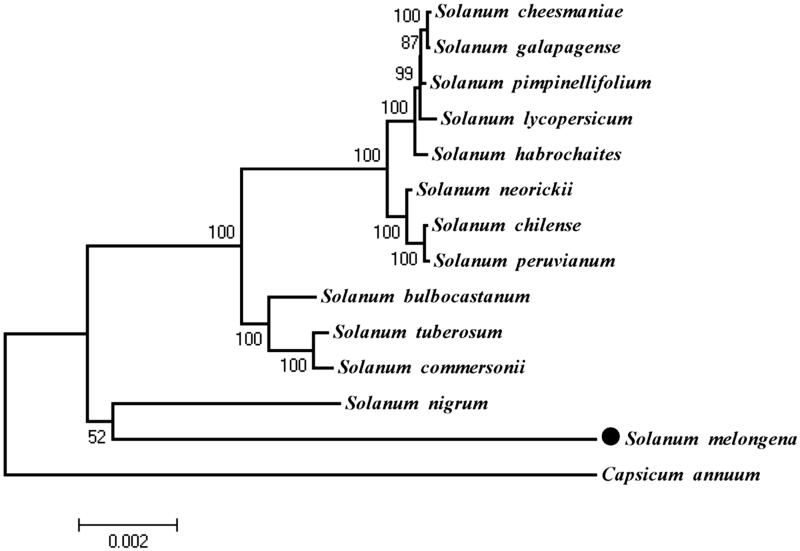
The phylogenetic analysis in this study included the 12 cp genomes of Solanum species (Chung et al. 2006; Daniell et al. 2006; Wu 2015) and the newly determined cp genome of S. melongena, using Capsicum annuum (Raveendar et al. 2015) as outgroup. ML analysis yielded in a phylogenetic tree with most of the nodes (8/11) supported by a bootstrap value of 100, and S. melongena clustered with S. nigrum supported by a bootstrap value of 52 (Figure 1), which was concordant with the previous studies in the genus Solanum (Wu 2015). The availability of cp genome of S. melongena provides valuable data for the future genetic improvement of this important vegetable species.
Figure 1.
Maximum-likelihood (ML) phylogenetic tree of Solanum melongena and the other 12 Solanum species using whole chloroplast genome sequences. Capsicum annuum was served as outgroup. The numbers on each node show the ML bootstrap support values. The accession numbers are as below: Solanum bulbocastanum (NC_007943), Solanum cheesmaniae (NC_026876), Solanum chilense (NC_026877), Solanum commersonii (NC_028069), Solanum galapagense (NC_026878), Solanum habrochaites (NC_026879), Solanum lycopersicum (NC_007898), Solanum neorickii (NC_026880), Solanum nigrum (NC_028070), Solanum peruvianum (NC_026881), Solanum pimpinellifolium (NC_026882), Solanum tuberosum (NC_008096), Capsicum annuum (NC_018552).
Disclosure statement
The authors declare that there are no conflicts of interest and are responsible for the content.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Azuma K, Ohyama A, Ippoushi K, Ichiyanagi T, Takeuchi A, Saito T, Fukuoka H.. 2008. Structures and antioxidant activity of anthocyanins in many accessions of eggplant and its related species. J Agric Food Chem. 56:10154–10159. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Jung JD, Park HW, Kim JH, Cha HW, Min SR, Jeong WJ, Liu JR.. 2006. The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep. 25:1369–1379. [DOI] [PubMed] [Google Scholar]
- Daunay MC, Lester RN.. 1988. The usefulness of taxonomy or Solanaceae breeders, with special reference to the genus Solanum and to Solanum melongena L. (eggplant). Capsicum Newslett. 7:70–79. [Google Scholar]
- Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK.. 2006. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 112:1503–1518. [DOI] [PubMed] [Google Scholar]
- Hirakawa H, Shirasawa K, Miyatake K, Nunome T, Negoro S, Ohyama A, Yamaguchi H, Isobe S, Tabata S, et al. 2014. Draft genome sequence of eggplant (Solanum melongena L.): the representative Solanum species indigenous to the old world. DNA Res. 21:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Park CE, Park GS, Seo YJ, So JH, Shin JH.. 2015. The whole chloroplast genome sequence of black nightshade plant (Solanum nigrum). Mitochondrial DNA. 28:1–2. [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J.. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 25:1966–1967. [DOI] [PubMed] [Google Scholar]
- Raveendar S, Na YW, Lee JR, Shim D, Ma KH, Lee SY, Chung JW.. 2015. The complete chloroplast genome of Capsicum annuum var. glabriusculum using Illumina sequencing. Molecules. 20:13080–13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Yoshida T, Monma S, Matsunaga H, Sato T, Saito A, Yamada T.. 2009. Development of the parthenocarpic eggplant cultivar ‘Anominori’. Jpn Agri Res Quart. 43:123–127. [Google Scholar]
- Sakata Y, Monma S, Narikawa T, Komochi S.. 1996. Evaluation of resistance to bacterial wilt and Verticillium wilt in eggplants (Solanum melongena L.) collected in Malaysia. J Jpn Soc Hort Sci. 65:81–88. [Google Scholar]
- Stommel JR, Whitaker BD.. 2003. Phenolic acid content and composition of eggplant fruit in a germplasm core subset. J Am Soc Hort Sci. 128:704–710. [Google Scholar]
- Wu Z. 2015. The completed eight chloroplast genomes of tomato from Solanum genus. Mitochondrial DNA. 21:1–3. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]