Abstract
Fragaria orientalis (Rosales: Rosaceae) as drought-resisting plant is distributed throughout northeast and central Asia. The complete chloroplast genome sequence of Fragaria orientalis is reported and characterized in this study. The genome size of F. orientalis is 147,835 bp, containing a pair of inverted repeats (IR) regions of 51,216 bp, which were separated by a large single copy (LSC) region of 83,232 bp and a small single copy (SSC) region of 13,387 bp. The genome contains 128 genes, including 84 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. A total of 17 of these genes are duplicated in the IR regions, 16 genes contain one intron, and ycf3 has two introns. Maximum likelihood phylogenetic analysis based on chloroplast genome sequences indicates that F. orientalis is closely related to Fragaria chiloensis and Fragaria virginiana from the family Rosaceae.
Keywords: Fragaria orientalis, chloroplast genome, phylogenetic analysis
Fragaria orientalis were found in 1934, tetraploid, had 28 chromosomes (Fedorova 1934). F. orientalis, belonging to the Rosaceae family, is native to northeast and central Asia (Guan et al. 2004). In order to adapt the environment, F. orientalis has evolved the features that are resistant to cold and drought (Guo et al. 2015). F. orientalis is a good wild germplasm resource of strawberry breeding (Gu et al. 2010). As a member of strawberry, F. orientalis is usually propagated by the stolon, so the fruit is small, and yield is low (Gu et al. 2010; Sun et al. 2010). F. orientalis tastes like bubblegum which is popular with Europe (http://www.dailymail.co.uk/sciencetech/article-2616658).
Based on the widely distribution, F. orientalis may have more genetic diversity than the species with narrow distribution. We will find the new taste strawberry by developing the genetic diversity of F. orientalis. So, our study had got the completion of chloroplast genome of F. orientalis, it will not only enhance the conservation efforts of this plant species, but also provide the plentiful gene resources for Fragaria breeding.
The materials of F. orientalis come from the NCGR-Corvallis (National Center for Genome Resources); the sequence platform is Illumina HiSeq1000, and the total bases are about 106 GB (Hirakawa et al. 2014). In the study, we use the raw data of F. orientalis (Hirakawa et al. 2014) to get the complete chloroplast genome sequence (GenBank: KY769126). We used the CLC Genomics Workbench v. 3.6.1 (CLC Inc., Aarhus, Denmark) to filter and assemble contigs. The circular map was obtained using the online program OGDRAW (Lohse et al. 2013).
The F. orientalis chloroplast genome is 147,835 bp long, with a typical circular structure, containing a pair of inverted repeat (IR) regions of 25,608 bp, which were separated by a large single copy (LSC) region of 83,232 bp and a small single copy (SSC) region of 13,387 bp. The chloroplast genome contains 128 genes, including 84 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Most of these genes occur as a single copy, however, seven protein-coding genes, seven tRNA genes, and four rRNA genes are duplicated in the IR regions. A total of 17 of these genes are duplicated in the IR regions, 16 genes contain one intron, especially, the gene ycf3 has two introns. The overall GC content of the chloroplast genome is 37.60% and the corresponding values in LSC, SSC, and IR regions are 35.23%, 30.22%, and 42.86%, respectively.
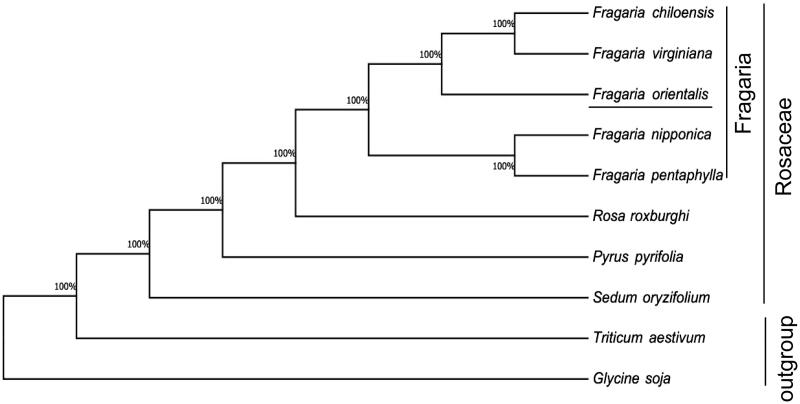
We constructed a maximum likelihood phylogenetic tree using seven species from the order Rosales (Figure 1), while Glycine soja and Triticum aestivum served as outgroup. All the chloroplast genome sequences were aligned using MEGA6 (Kumar et al. 2016). Our result showed that F. orientalis is most closely related to F. chiloensis and F. virginiana from the family Rosales. The complete chloroplast genome of F. orientalis would provide valuable information for biodiversity conservation and genetic breeding.
Figure 1.
Maximum likelihood phylogenetic tree of F. orientalis with seven other species in the Rosales based on complete chloroplast genome sequences, while using Glycine soja and Triticum aestivumas outgroup. Accession numbers are listed as below: Fragaria chiloensis (NC_019601), Fragaria nipponica (NC_035500), Fragaria pentaphylla (NC_034347), Fragaria virginiana (NC_019602), Pyrus pyrifolia (NC_015996), Rosa roxburghii (NC_032038), Sedum oryzifolium (NC_027837), Glycine soja (NC_022868.1), and Triticum aestivum (NC_002762).
Acknowledgements
We thank the editor and anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
There are no conflicts of interest to declare.
References
- Fedorova N. 1934. Polyploid inter-specific hybrids in the genus Fragaria. Genetica. 16:524–541. [Google Scholar]
- Gu DZ, Gao HD, Zhu JY, Jiang YT, Feng Y, Sun ZL, Liu XL.. 2010. Uniform design for in vitro culture and plant regeneration through petals of Fragaria orientalis Lozinsk varconcolor(Kitag) Liou et C.Y.Li. J Zhejiang Univ. 37:700–704. [Google Scholar]
- Guan W, Minsheng YE, Keming MA, Liu G, Wang X.. 2004. Vegetation classification and the main types of vegetation of the dry valley of Minjiang River. J Mountain Res. 22:679–686. [Google Scholar]
- Guo S, Zhu Y, Liu D.. 2015. Effect of drought stress on morphological and physiological characteristics of Fragaria orientalis. North Horticult.13:40–43. [Google Scholar]
- Hirakawa H, Shirasawa K, Kosugi S, Tashiro K, Nakayama S, Yamada M, Kohara M, Watanabe A, Kishida Y, Fujishiro T, et al. 2014. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 21:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZL, Di-Zhou GU, Zhu JY, Jiang YT, Feng Y, Yuan Y.. 2010. Study on the growth trait of Fragaria orientalis var. concolor by petal induction. Guangdong Agric Sci. 37:112–114. [Google Scholar]