Abstract
In the present study, we report the first complete mitochondrial DNA genome of the genus Callipogon based on C. relictus, a natural monument and endangered species in South Korea. The mitogenome is 15,742 base pairs with 13 protein coding genes (PCGs), two rRNAs, 22 tRNAs, and a 1033 bp long AT-rich region. The overall base composition was 67.3% AT and 32.7 GC. Among 13 PCGs, seven genes (Nad2, Atp8, Atp6, Nad4L, Nad6, Cob, Nad1) harbour the typical stop codon TAA or TAG, whereas remaining five genes terminate with T. Interestingly, Cox3 employs AGA as the termination codon.
Keywords: Korean natural monument, endangered species, mitogenome, longhorn beetle, Callipogon relictus
Callipogon is a genus with a disjunct distribution: one species, C. relictus, in Northeast Asia and seven in Central America (Cherepanov 1988; Lim, Kim, et al. 2013; Lim, Jung, et al. 2013). Callipogon relictus was designated as a Korean natural monument in 1968 and an endangered species in 2012 for a sharp decline of the population in Korea (Lim, Jung, et al. 2013).
A male and a female were found in Gwangneung forest, Korea in 2014 and 2015, respectively, and stored in absolute ETOH at −80 °C. The voucher specimens (accession no. NHC-A-2015-054, NHC-A-2016-120) are deposited in the Natural Heritage Center, Korea. We extracted genomic DNA from the bodies with DNA extraction kit (Takara, Japan). The extracted DNA sample was analysed using Illumina MiSeq. A total of 24,202,986 reads were analysed to generate 3,654,650,886 base pairs of sequence and assembled in Geneious 10.0.4 (Kearse et al. 2012). A total of 50,128 reads were assembled with an average coverage of 478.4Χ. Phylogenetic analysis was performed based on the nucleotide sequences of the 13 PCGs. Secondary structure prediction of tRNAs except for two tRNAs (tRNAAsn and tRNASer), was done in Lowe Lab tRNAscan-SE Search Server (Schattner et al. 2005) and the secondary structure prediction of tRNAAsn in Bioinformatics Web Server for RNA (http://rtools.cbrc.jp).
The C. relictus complete mitogenome, 15,742 bp, has 37 individual genes (13 protein coding genes (PCGs), two ribosomal RNAs, 22 tRNAs, and a 1033 bp long A + T rich region). The overall base composition was 67.3% AT and 32.7% GC. The 13 PCGs are similar to other coleopteran insects, and 12 genes begin with typical ATN codon: four (Atp6, Cox3, Nad4L, Cob) start with ATG, three (Nad2, Nad3, Nad1) with ATT, two (Cox2, Nad6) with ATC and three (Atp8, Nad5, Nad4) with ATA. ACC was observed as an atypical start codon for Cox1. Among 13 PCGs, seven genes (Nad2, Atp8, Atp6, Nad4L, Nad6, Cob, Nad1) harbour the typical stop codon TAA or TAG, whereas remaining five genes terminate with T. Interestingly, Cox3 employs AGA as the termination codon. Taken together, one gene (Cox1) starts atypical codon, and 6 genes (Cox1, Cox2, Cox3, Nad3, Nad5, Nad4) possess atypical stop codon. The mitogenome of C. relictus has 22 tRNA genes: two isotypes for Leu and Ser, one type for the other 18 types of amino acid. With the exception of tRNAser, the dihydrouridine arm of which forms a simple loop, 21 tRNAs can be folded into the typical clover-leaf structure. While the order of tRNAAla and tRNAArg among the ARNSEF (Ala, Arg, Asn, Ser, Glu, Phe) cluster between the Nad3 and Nad5 genes is reversed in some chrysomelid beetles and aquatic elateriform beetles (Timmermans and Vogler 2012; Timmermans et al. 2015). The lengths of 16S and 12S rRNA genes are 1287 and 817 bp, respectively, which are well within the range of the respective genes detected in the coleopteran mitochondrial rRNAs (Kim et al. 2009; Chiu et al. 2016). A large non-coding AT-rich region (79.6%) of 1033 nucleotides is located between 12S rRNA and tRNAIle. Although certain repetitive sequences were not investigated in AT-rich region in C. relictus, a 16 bp-long poly-A, a 9 bp-long poly-T, and microsatellite-like TA repeats were scattered throughout this region (Table 1).
Table 1.
Mitochondrial genome of Callipogon relictus.
| Gene | Direction | Location | Length | Anticodon | Codon |
|
|---|---|---|---|---|---|---|
| Start | Stop | |||||
| tRNAIle | F | 1–67 | 67 | GAT (29–31) | ||
| tRNAGln | R | 70–140 | 71 | CAA (108–110) | ||
| tRNAMet | F | 143–211 | 69 | CAT (173–175) | ||
| Nad2 | F | 212–1225 | 1014 | ATT | TAA | |
| tRNATrp | F | 1224–1291 | 68 | TCA (1258–1260) | ||
| tRNACys | R | 1292–1357 | 66 | GCA (1326–1328) | ||
| tRNATyr | R | 1358–1422 | 65 | GTA (1391–1393) | ||
| Cox1 | F | 1415–2957 | 1513 | ACC | T | |
| tRNALeu | F | 2958–3022 | 65 | TAA (2987–2989) | ||
| Cox2 | F | 3023–3707 | 685 | ATC | T | |
| tRNALys | F | 3708–3777 | 70 | TTT (3737–3739) | ||
| tRNAAsp | F | 3778–3844 | 67 | GTC (3809–3811) | ||
| Atp8 | F | 3845–4000 | 156 | ATA | TAA | |
| Atp6 | F | 3994–4665 | 672 | ATG | TAA | |
| Cox3 | F | 4665–5453 | 786 | ATG | AGA | |
| tRNAGly | F | 5451–5515 | 65 | TCC (5482–5484) | ||
| Nad3 | F | 5516–5867 | 352 | ATT | T | |
| tRNAAla | F | 5868–5933 | 66 | TGC (5897–5899) | ||
| tRNAArg | F | 5930–5998 | 69 | TCG (5962–5964) | ||
| tRNAAsn | F | 5999–6064 | 66 | GTT (6031–6033) | ||
| tRNASer | F | 6064–6131 | 68 | TCT (6096–6098) | ||
| tRNAGlu | F | 6132–6194 | 63 | TTC (6162–6164) | ||
| tRNAPhe | R | 6193–6255 | 63 | GAA (6223–6225) | ||
| Nad5 | R | 6256–7978 | 1723 | ATA | T | |
| tRNAHis | R | 7976–8040 | 65 | GTG (8007–8009) | ||
| Nad4 | R | 8041–9370 | 1330 | ATA | T | |
| Nad4L | R | 9367–9651 | 285 | ATG | TAA | |
| tRNAThr | F | 9654–9717 | 64 | TGT (9684–9686) | ||
| tRNAPro | R | 9718–9782 | 65 | TGG (9750–9752) | ||
| Nad6 | F | 9784–10,299 | 516 | ATC | TAA | |
| Cob | F | 10,299–11,438 | 1140 | ATG | TAG | |
| tRNASer | F | 11,437–11,503 | 67 | TGA (11,466–11,468) | ||
| Nad1 | R | 11,521–12,465 | 945 | ATT | TAG | |
| tRNALeu | R | 12,472–12,536 | 65 | TAG (12,505–12,507) | ||
| 16S rRNA | R | 12,538–13,824 | 1287 | |||
| tRNAVal | R | 13,825–13,892 | 68 | TAC (13,861–13,863) | ||
| 12S rRNA | R | 13,893–14,709 | 817 | |||
| A + T rich region | R | 14,710–15,742 | 1033 | |||
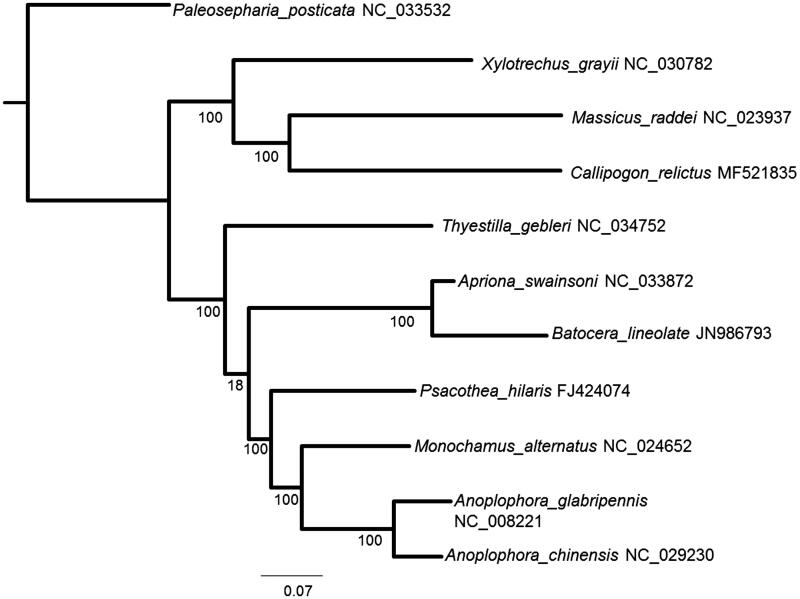
A phylogenetic position of C. relictus was inferred, and the analysis clearly showed the beetle as a distinct species among the other cerambycid species (Figure 1).
Figure 1.
A maximum-likelihood tree inferred from the nucleotide sequences of 13 PCGs in the mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values. Paleosepharia posticata was used as an outgroup. Alphanumeric terms indicate the GenBank accession numbers.
Prior to the initiation of a conservation breeding program, the complete mitogenome of C. relictus from this study will be used to determine the population genetic structure and diversity among the species populations from all three countries.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of Callipogon relictus has been assigned GenBank accession number MF521835.
Acknowledgements
We thank Drs Sang-Yong Kim (Korea National Arboretum, Yangpyeong, Korea), Bong-Woo Lee (Korea Forest Service, Daejeon, Korea) and Mr. Ik-Je Choi (Gyeongsangbuk-do Foundation Original Seed Production Center, Sangju, Korea) for their kind assistance in the field, Gwangneung forest, Korea.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. No potential conflict of interest was reported by the authors.
References
- Cherepanov AI. 1988. Cerambycidae of Northern Asia. Vol. 1. Prioninae, disteniinae, lepturinae, aseminae. New Delhi: Amerind Publishing.
- Chiu WC, Yeh WB, Chen ME, Yang MM.. 2016. Complete mitochondrial genome of Aeolesthes oenochrous (Fairmaire) (Coleoptera: Cerambycidae): an endangered and colorful longhorn beetle. Mitochondrial DNA A DNA Mapp Seq Anal. 27:686–687. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious Basic: an integrated and extendable desk to software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KG, Hong MY, Kim MJ, Im HH, Kim MI, Bae CH, Seo SJ, Lee SH, Kim I.. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol Cells. 27:429–441. [DOI] [PubMed] [Google Scholar]
- Lim J, Jung SY, Lim JS, Jang J, Kim KM, Lee YM, Lee BW.. 2013. A review of host plants of Cerambycidae (Coleoptera: Chrysomeloidea) with new host records for fourteen cerambycids, including the Asian longhorn beetle (Anoplophora glabrippenis Motschulsky), in Korea. Korean J Appl Entomol. 53:111–133. [Google Scholar]
- Lim J, Kim M, Kim IK, Jung S, Lim JS, Park SY, Kim KM, Byun BK, Lee BW, Lee S.. 2013. Molecular identification and larval description of Callipogon relictus Semenov (Coleoptera: Cerambycidae), a natural monument of South Korea. J Asia Pac Entomol. 16:223–227. [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJTN, Barton C, Haran J, Ahrens D, Culverwell CL, Ollikainen A, Dodsworth S, Foster PG, Bocak L, Vogler P.. 2015. Family-level sampling of mitochondrial genomes in Coleoptera: compositional heterogeneity and phylogenetics. Genome Biol Evol. 8:161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJTN, Vogler AP.. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63:299–304. [DOI] [PubMed] [Google Scholar]