Abstract
In order to supply genetic information of Camellia sinensis cultivar Anhua, characterization of the complete chloroplast genome sequence was reported based on high-throughput sequencing data. The complete cp genome of C. sinensis cultivar Anhua is shorter than other C. sinensis cultivars with 157,025 bp in length, comprising a large single copy (LSC) region of 86,585 bp and a small single copy (SSC) region of 18,276 bp, separated by two inverted repeat regions (IRs) of 26,082 bp. The overall G + C content is 37.30%. The genome contained total of 135 genes, including 90 protein coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed all the cultivars were clustered into a group except C. sinensis var. pubilimba.
Keywords: Chloroplast genome, Camellia sinensis, dark tea, Illumina, Sequencing
Camellia sinensis cultivar Anhua, which belongs to Theaceae, is evergreen small tree or shrub. Leaf of the economic plant is the major source of dark tea, which becomes increasingly popular owing to its special health benefits. In accordance with dark tea production area, C. sinensis is cultivated mainly in Hunan, Sichuan, and Yunnan provinces in China (Zheng et al. 2015). The quality of dark tea have close relationship with cultivar of C. sinensis and geographical teas are formed (Ning et al. 2016). Because of the special planting environment, C. sinensis cultivar Anhua is famous and Anhua brick dark tea sell well (Chen et al. 2015). Except for materials of dark tea, other economic functions are explored based on the rich secondary metabolites (Shen et al. 2018). Tea cultivation has a long history and there are so many cultivar populations. Genetic diversity of C. sinensis cultivars were revealed in many researches (Roy and Chakraborty 2009; Yuan et al. 2015). Lots of germplasm resources of C. sinensis were lost gradually due to the long period artificial selection. Thus, it is urgent and necessary to protect and make use of C. sinensis, the germplasm resources. In this study, characterization of the complete chloroplast genome sequence of C. sinensis cultivar Anhua was reported based on high-throughput sequencing data with the accession number MH042531.
Fresh leaves of C. sinensis cultivar Anhua were collected from an individual in plant garden of Hunan City university (Hunan, China; N28°32′35.76″, E112°22′53.53″) and the samples were kept in the laboratory at −80 °C under the accession number 20171121DM. The whole genomic DNA was extracted from fresh leaves, using DNeasy Plant Mini Kit (QIAGEN,Hilden, Germany).Total chloroplast DNA was extracted with Plant Chloroplast Purification Kit (BTN120308, Beijing, China) and Column Plant DNA Extraction Kit(K183001, Massachusetts, USA) and sequenced by Illumina Miseq Platform (California, USA). Filtration and annotation of the whole genome and drawing of a circular map referred to Zhang’s method by using Trimmomatic v0.32, DOGMA and cpGAVAS, OGDRAW (Wyman et al. 2004; Lohse et al. 2007; Yong et al. 2012; Bolger et al. 2014; Zhang et al. 2017).
The complete cp genome of C. sinensis cultivar Anhua is shorter than other C. sinensis cultivars (KJ806281, KJ806279, KC143082, NC020019, KJ996106, KJ806280, KF562708) with 157,025 bp in length, comprising of a large single copy (LSC) region of 86,585 bp and a small single copy (SSC) region of 18,276 bp, separated by two inverted repeat regions (IRs) of 26,082 bp. The overall G + C content is 37.30%. The genome contained total of 135 genes, including 90 protein coding genes, 37 tRNA genes, and 8 rRNA genes. Among the protein coding genes, atpF, rpoC1, rpl2, ndhB, ndhA, ycf1 contained one intron, ycf3 and clpP included a couple of introns.
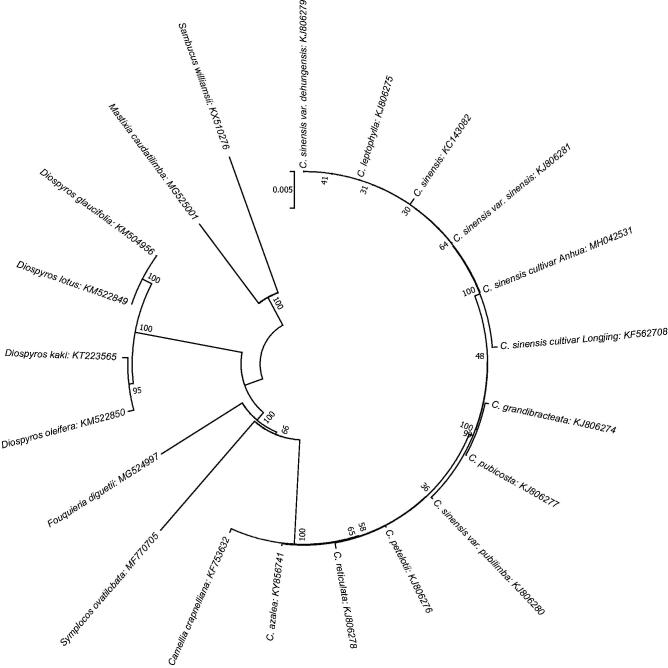
A molecular phylogenetic tree was constructed with the MEGA7.0 to confirm phylogenetic position of C. sinensis cultivar Anhua (Kumar et al. 2016) based on all common protein coding genes of other cultivars and species from different genus (Figure 1). The result showed all the cultivars were clustered into a group except C. sinensis var. pubilimba. A good foundation for population genomic studies and genetic engineering was laid through this annotated chloroplast genome.
Figure 1.
The phylogenetic tree based on 21 complete chloroplast genome sequences. The neighbour-joining (NJ) phylogenetic tree was constructed with MEGA 7 (with 1000 bootstrap replicates).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Deng Y, Gong Z, Zhu H, University HA.. 2015. Analysis of the evolution of technological innovation model of Anhua dark tea industry. J Agric. 5:96–101. [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R.. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Ning J, Fang J, Luo X, Zhang Z, Wan X.. 2016. Identification of dark tea (Camellia sinensis (L.)) origins according to chemical composition combined with Bayes Classification pattern recognition. Adv J Food SciTechnol. 12:150–154. [Google Scholar]
- Roy SC, Chakraborty BN.. 2009. Genetic diversity and relationships among tea (Camellia sinensis) cultivars as revealed by RAPD and ISSR based fingerprinting. Indian J Biotechnol. 8:370–376. [Google Scholar]
- Shen J, Zou Z, Zhang X, Zhou L, Wang Y, Fang W, Zhu X.. 2018. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hort Res. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yong Y, Zheng X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Xiong LG, Deng TT, Wu Y, Li J, Liu SQ, Huang JA, Liu ZH.. 2015. Comparative profiling of gene expression in Camellia sinensis L. cultivar AnJiBaiCha leaves during periodic albinism. Gene. 561:23. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Yang G, Tang Y, Xu Z.. 2017. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondrial DNA B. 2:843–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng WJ, Wan XC, Bao GH.. 2015. Brick dark tea: a review of the manufacture, chemical constituents and bioconversion of the major chemical components during fermentation. Phytochem Rev. 14:499–523. [Google Scholar]