Abstract
The long-horned caddisfly Triaenodes tardus Milne, 1934 (Leptoceridae), is a widespread herbivorous North American caddisfly found in both lentic and lotic habitats. Whole genome Illumina sequencing allowed the assembly of a complete circular mitogenome of 14,963 bp from T. tardus consisting of 73.4% AT nucleotides, 22 tRNAs, 13 protein-coding genes, two rRNAs and a control region in the ancestral insect gene order. Triaenodes tardus COX1 features an atypical TTG start codon as in some lepdioptera and prokaryotes. Phylogenetic reconstruction places T. tardus as sister to Sericostoma personatum (Sericostomatidae) within a monophyletic Order Trichoptera, which is consistent with previous phylogenetic hypotheses.
Keywords: Illumina sequencing, mitogenomics, Trichoptera, Leptoceridae, TTG initiation codon
Most caddisfly larvae (Insecta: Trichoptera) are benthic aquatic detritivores. However, caddisflies in family Leptoceridae have morphological adaptations for swimming and species in leptocerid genus Triaenodes are herbivorous (Gall et al. 2011). Triaenodes tardus Milne, 1934 is a widespread North American species (NatureServe 2017) whose larvae are found in littoral vegetation in lotic and slowly flowing lentic habitats (Schwiebert 2007). Here we present the first complete mitogenome for family Leptoceridae from T. tardus.
On 14–15 August 2015, a USDA blacklight trap (Winter 2000) was deployed to collect night-flying insects at the Living Prairie Museum (GPS 49.889607 N, −97.270487 W), 12.9 hectares of relict prairie in Winnipeg, Manitoba, Canada (Living Prairie Mitogenomics Consortium 2017). Two adult T. tardus were trapped (specimens: 2015.08.14.063, 2015.08.14.077; determined by morphology and COX1 barcodes). Specimen 2015.08.14.077 was pinned and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher JBWM0361497).
DNA was prepared (McCullagh and Marcus 2015) and sequenced by Illumina MiSeq (San Diego, CA) (Peters and Marcus 2017). The mitogenome of T. tardus (Genbank MG201852) was assembled by Geneious 10.1.2 from 8,257,770 paired 75 bp reads using an Anabolia bimaculata (Trichoptera: Limnephilidae) reference mitogenome (MF680449) (Peirson and Marcus 2017). Annotation was in reference to A. bimaculata and Sericostoma personatum (Trichoptera: Sericostomatidae, KP455290) mitogenomes (Dietz et al. 2015). The T. tardus nuclear rRNA repeat (Genbank MG201853) was also assembled and annotated using A. bimaculata (MF680448) (Peirson and Marcus 2017) and Stenopsyche marmorata (Trichoptera: Stenopsychidae, LC094265.1) reference sequences.
The T. tardus circular 14,963 bp mitogenome assembly was composed of 6,952 paired reads with nucleotide composition: 33.2% A, 13.8% C, 12.9% G, and 40.2% T. The gene composition and order in T. tardus is identical to all known trichopteran mitogenomes except for Hydropsyche pellucidula (Hydropsychidae) (Linard et al. 2017). Triaenodes tardus COX1 features an atypical TTG start codon as in some Lepidoptera (Chen et al. 2012) and prokaryotes (Asano 2014). The mitogenome contains three protein-coding genes (COX2, NAD4, and NAD5) with single-nucleotide (T) stop codons, and two protein-coding genes (ATP6 and NAD3) with two-nucleotide (TA) stop codons completed by post-transcriptional addition of 3′ A residues. All tRNAs have typical cloverleaf secondary structures except for trnS (AGN) where the dihydrouridine arm is replaced by a loop as determined in Mfold (Zuker 2003). The rRNAs and control region are typical for Trichoptera (Peirson and Marcus 2017).
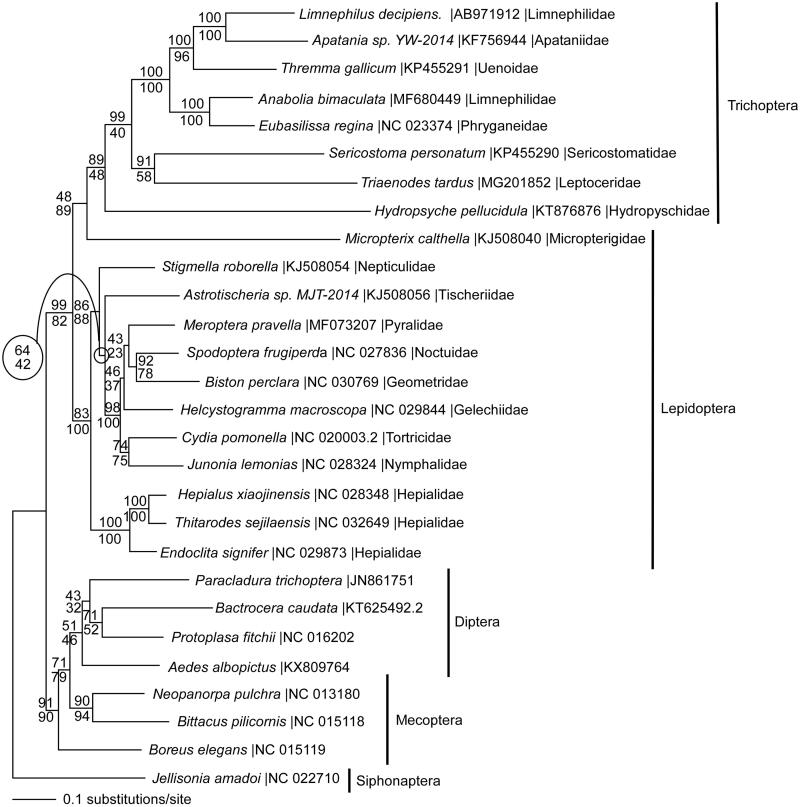
We reconstructed a phylogeny using 13 mitochondrial protein coding genes from T. tardus, seven other trichopteran species, and species in related holometabolous insect orders. Each gene was aligned in CLUSTAL Omega (Sievers et al. 2011), concatenated, and analyzed by maximum likelihood (ML) and parsimony in PAUP* 4.0b8/4.0d78 (Swofford 2002) (Figure 1). ML phylogenetic analysis shows Trichoptera as monophyletic; places T. tardus as sister to Sericostoma personatum, consistent with previous phylogenetic hypotheses (Kjer et al. 2002); and the primitive lepidopteran Micropterix calthella (Micropterigidae) was found to be sister to the Trichoptera, while the Trichoptera + Micropterix clade was sister to the remaining Lepidoptera (Peirson and Marcus 2017). The trichopteran family Limnephilidae is not monophyletic in this analysis, which warrants further investigation.
Figure 1.
Maximum likelihood phylogeny (GTR + I + G model, I = 0.1960, G = 0.6200, likelihood score 175,277.12756) of Triaenodes tardus and other Trichoptera species with representatives from related insect orders Lepidoptera (moths and butterflies), Diptera (flies), Mecoptera (scorpionfiles), and Siphonaptera (fleas) based on 1 million random addition heuristic search replicates (with tree bisection and reconnection) of mitochondrial protein coding genes. One million maximum parsimony heuristic search replicates produced a single nearly identical tree (42,585 steps) except that Micropterix is the sister taxon to Hydropsyche, rather than to the entire trichopteran clade. Maximum likelihood bootstrap values are above nodes and maximum parsimony bootstrap values are below nodes (each from 1 million random fast addition search replicates).
Acknowledgements
The authors thank Sarah Semmler and Kyle Lucyk for permitting and encouraging our work at the Living Prairie Museum. The authors thank Melissa Peters for help with fieldwork and Aleksandar Ilik and Debbie Tsuyuki (Children’s Hospital Research Institute of Manitoba Next Generation Sequencing Platform) for assistance with library preparation and sequencing.
Disclosure statement
The authors report no conflicts of interest, and are solely responsible for this paper.
References
- Asano K. 2014. Why is start codon selection so precise in eukaryotes? Translation (Austin). 2:e28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Li-Li T, Shi Q-H, Cao T-W, Hao J-S.. 2012. Complete mitogenome of the Lesser Purple Emperor Apatura ilia (Lepidoptera: Nymphalidae: Apaturinae) and comparison with other nymphalid butterflies. Zool Res. 33:191–201. [DOI] [PubMed] [Google Scholar]
- Dietz L, Brand P, Eschner LM, Leese F.. 2015. The mitochondrial genomes of the caddisflies Sericostoma personatum and Thremma gallicum (Insecta: Trichoptera). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3293–3294. [DOI] [PubMed] [Google Scholar]
- Gall BG, Hopkins GR, Brodie ED Jr.. 2011. Mechanics and ecological role of swimming behavior in the caddisfly larvae Triaenodes tardus. J Insect Behav. 24:317–328. [Google Scholar]
- Kjer KM, Blahnik RJ, Holzenthal RW.. 2002. Phylogeny of caddisflies (Insecta, Trichoptera). Zool Scr. 31:83–91. [Google Scholar]
- Linard B, Arribas P, Andujar C, Crampton-Platt A, Vogler AP.. 2017. The mitogenome of Hydropsyche pellucidula (Hydropsychidae): first gene arrangement in the insect order Trichoptera. Mitochondrial DNA A DNA Mapp Seq Anal. 28:71–72. [DOI] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA B Resour. 2:344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh BS, Marcus JM.. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18:749–755. [Google Scholar]
- NatureServe. (2017). NatureServe Explorer: an online encyclopedia of life [web application]. Version 7.1. NatureServe, Arlington, VI; Available from: http://explorer.natureserve.org [Google Scholar]
- Peirson DSJ, Marcus JM.. 2017. The complete mitochondrial genome of the North American caddisfly Anabolia bimaculata (Insecta: Trichoptera: Limnephilidae). Mitochondrial DNA B Resour. 2:595–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Marcus JM.. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Ent. 42:288–300. [Google Scholar]
- Schwiebert E. 2007. Nymphs, stoneflies, caddisflies, and other important insects: including the lesser mayflies. Volume II Boston (MA): Lyons Press. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Version 4. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Winter WD. 2000. Basic techniques for observing and studying moths and butterflies. Vol 5 Los Angeles (CA): The Lepidopterists' Society. [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]