Abstract
Macadamia ternifolia is a subtropical fruit tree of the family Proteaceae. Chloroplast genome sequences play a significant role in the development of molecular markers in plant phylogenetic and population genetic studies. In this study, we report the complete chloroplast genome sequence of M. ternifolia. The chloroplast genome is 159,669 bp long and includes 113 genes. Its LSC, SSC, and IR regions are 88,072, 18,801, and 26,408 bp long, respectively. Sequence comparison of M. ternifolia and M. integrifolia indicates large sequence conservation between these two species, only few sequence variations including single nucleotide polymorphisms (SNPs) and inserts/deletes (InDels) were detected.
Keywords: Chloroplast genome, Macadamia ternifolia, Proteaceae
Macadamia (Macadamia spp.) is a subtropical fruit tree from Australia and is widely cultivated in many other countries for the high oil content of its edible kernel (Mason and McConachie, 1994). There are three major fruit tree species in the genus Macadamia: M. integrifolia, M. tetraphylla, and M. ternifolia (Aradhya et al. 1998; Hardner et al. 2009), and M. ternifolia is the wild relative of the other two species. Chloroplast genome sequences can provide useful molecular markers for plant genetic studies. A previous study has reported the complete chloroplast genome sequence of M. integrifolia (Nock et al. 2014). In this study, we report the complete chloroplast genome of M. ternifolia, and hope it would aid further phylogenetic and population genetic studies of the Macadamia species.
DNA material was isolated from mature leaves of a M. ternifolia plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China by using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). A specimen of this tree was conserved in YITC. About 10 μg isolated DNA was sent to BGI Shenzhen for library construction and genome sequencing on the Illumina Hiseq 2000 Platform. After genome sequencing, a total of 3.1 Gbp reads in fastq format were obtained and subjected to chloroplast genome assembly. The complete chloroplast genome was annotated with Dual Organelle GenoMe Annotator (DOGMA; Wyman et al. 2004) and submitted to the Genbank (http://www.ncbi.nlm.nih.gov/) under the accession number of MF678834. A physical map of the chloroplast genome was generated by OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. 2013).
Our assembly of the M. ternifolia resulted in a final sequence of 159,669 bp in length with no gap. The overall A-T content of the chloroplast genome was 61.2%. This chloroplast genome included a typical quadripartite structure with the large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions of 88,072, 18,801, and 26,408 bp long, respectively. Genome annotation showed 113 full length genes including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The genome organization, gene content, and gene relative positions were almost identical to the previously reported M. integrifolia chloroplast genome (Nock et al. 2014). Eighteen genes were duplicated in the IR regions. Fifteen genes contained one intron, while three had two introns. Sequence comparison of M. ternifolia and M. integrifolia indicated large sequence conservation between these two species, only few sequence variations of single nucleotide polymorphisms (SNPs) and inserts/deletes (InDels) were detected.
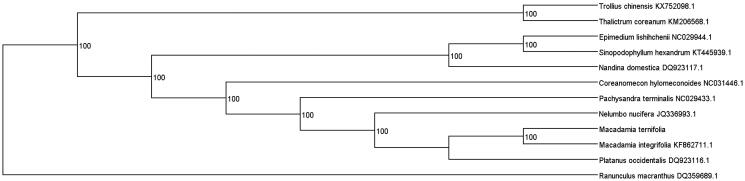
Phylogenetic tree included M. ternifolia, M. integrifolia, and the other 11 representative early diverging eudicotyledons species was constructed. Maximum-likelihood (ML) analysis exhibited that M. integrifolia clustered with M. ternifolia, and other species were highly supported by a bootstrap value of 100 (Figure 1). The evolutionary relationships of these analysed species are consistent with previously reported results (Nock et al. 2014).
Figure 1.
Maximum-likelihood (ML) phylogenetic tree of M. integrifoia and the other 11 representative early diverging eudicotyledons species. Number above each node indicates the ML bootstrap support values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Aradhya MK, Yee LK, Zee FT, Manshardt RM.. 1998. Genetic variability in Macadamia. Genet Resour Crop Ev. 45:19–32. [Google Scholar]
- Hardner CM, Peace C, Lowe AJ, Neal J, Pisanu P, Powell M, Schmidt A, Spain C, Williams K.. 2009. Genetic resources and domestication of Macadamia. Hortic Rev. 35:1–125. [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. Organellar GenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R, McConachie I.. 1994. A hard nut to crack: a review of the Australian Macadamia nut industry. Food Australia. 46:466. [Google Scholar]
- Nock CJ, Baten A, King GJ.. 2014. Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC Genomics. 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]